|
SCL
1.0
Standard Control Library : Control, dynamics, physics, and simulation
|
|
SCL
1.0
Standard Control Library : Control, dynamics, physics, and simulation
|
#include <CActuatorMuscle.hpp>
Public Member Functions | |
| virtual sBool | computeJacobian (const Eigen::VectorXd &arg_q, Eigen::VectorXd &ret_J) |
| std::string | getName () const |
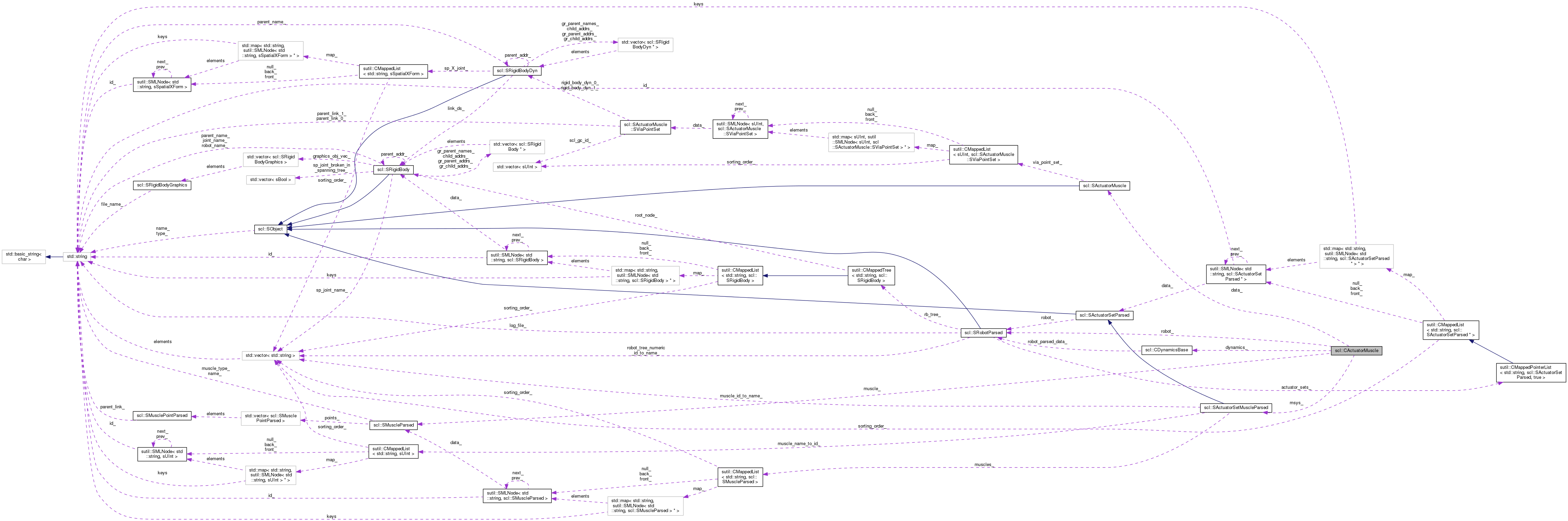
| virtual sBool | init (const std::string &arg_name, const SRobotParsed *arg_robot, const SActuatorSetMuscleParsed *arg_msys, const sutil::CMappedList< std::string, SRigidBodyDyn > &arg_rbdtree, CDynamicsBase *arg_dynamics) |
| virtual sBool | hasBeenInit () |
| CActuatorMuscle () | |
| virtual | ~CActuatorMuscle () |
Protected Attributes | |
| SActuatorMuscle | data_ |
| const SRobotParsed * | robot_ |
| const SActuatorSetMuscleParsed * | msys_ |
| const SMuscleParsed * | muscle_ |
| CDynamicsBase * | dynamics_ |
Models a muscle as a linear actuator with zero force generation delay and a stiff tendon, which eliminates slack.
Muscles may be connected to one or more limbs through multiple via-points.
The muscle length is determined by the distance between via-points that cross one or more generalized coordinates. Since the other via-points are fixed to a surface, they don't contribute to the overall length. This allows a simpler formulation of the muscle length gradient (Jacobian) matrix. Computing the model pair-wise between gc crossing via-points reduces the worst case complexity to O(n*2*k), where k are the gcs spanned by successive via-points.
NOTE : This doesn't accommodate muscle via points that slide across surfaces. While those are more biologically realistic, they involve solving a shortest path formulation over a curved surface (the Brachistochrone problem). The sliding point might be better for large motions across complex joint complexes.
| scl::CActuatorMuscle::CActuatorMuscle | ( | ) |
Default constructor. Sets stuff to NULL.
|
inlinevirtual |
Default destructor. Does nothing
|
virtual |
Computes the contribution of a single muscle to the entire Jacobian.
Positive muscle force => Muscle contraction Negative muscle force => Muscle pushes (usually an error)
Note for modeling biological musculoskeletal systems: Formulation is on a gc-by-gc basis. Ie. The muscle is considered piece- wise linear and each piece's contribution is measured independently. This is an approximation, but more detailed continuous models are hard to validate experimentally. Moreover, given the variability introduced by canonical musculoskeletal scaling, the errors introduced by piece- wise length gradients are probably within the envelope of experimental error. This demands biological investigation on a subject-by-subject basis.
To analytically compute the Jacobian, we use the following method:
δl = J . δq || l = muscle len. q = gen coord. J = row in Jm δli = J(i) . δq(i) || δli = change across muscle's i'th spanned gc li = |x_fixed + x_pre_gc - (x_fixed+xpost_gc)| || Part of muscle doesn't move. || = L2 norm δli/δq(i) = δ(|x_pre_gc - xpost_gc|)/δq(i) || Gradient wrt. q(i) = Gradient of path length between two attachment points (p0 and p1) = δ/δq ( |p0 - p1| ) = δ/δq ( sqrt( (p0-p1)' * (p0-p1) ) ) = 1/(2 * sqrt( (p0-p1)' * (p0-p1) ) ) * δ/δq ( (p0-p1)' * (p0-p1) ) = 1/(2 * sqrt( (p0-p1)' * (p0-p1) ) ) * 2* (p0-p1)' δ/δq (p0-p1) = 1/sqrt( (p0-p1)' * (p0-p1) ) * (p0-p1)' ( δ/δq (p0) - δ/δq (p1) ) = 1/d.norm() * d' ( δ/δq (p0) - δ/δq (p1) ); where d = p0 - p1
J_muscle_i (row-vector) = Sum_over_attachment_point_pairs_di [ |d|^-1 * d' (J_p0 - J_p1) ]
NOTE TODO : This can be greatly improved by computing Jacobians wrt. the parent link instead of wrt. the root node. Massive performance hit. NOTE TODO : Need to dig into the dynamics engine implementation and enable Jacobians wrt. some arbit link, and Jacobians with local xyz offset wrt node.
Some actuator sets don't directly actuate the generalized coordinates and require a Jacobian to compute their contribution to the generalized forces.
Each actuator instance must implement this.
| arg_q | The generalized coordinates |
| ret_J | The Jacobian for this muscle del.length = J * del.q |
NOTE : The Jacobian is a row vector with non-zero entries only at the spanned generalized coordinates
|
inlinevirtual |
Has this actuator been initialized
|
virtual |
Initializes the actuator. This involves determining whether the muscle matches the robot etc. It also sets up the Jacobians to be computed etc.
|
protected |
The data struct for the muscle actuator
|
protected |
Dynamics specification (to compute robot Jacobians)
|
protected |
The parsed muscle specification
|
protected |
The muscle in the system
|
protected |
The parent robot to which this actuator is attached
 1.8.6
1.8.6