Our Research:
| Cells, like whole organisms, have an incredible diversity of form, but unlike whole organisms, we know significantly less about how cells develop. Patterning at the cellular level allows cells to carry out specialized functions. In the Feldman Lab, we are interested in understanding the mechanisms underlying cell patterning. In particular, we are currently focused on understanding how microtubules become spatially organized, a key aspect of differentiation across different tissues and organisms. In addition, we study fundamental aspects of cell polarization, focusing on symmetry breaking cues that establish polarity in epithelia. We generally ask mechanistic cell biological questions in a living, developing organism using a genetic, biochemical, and live imaging approach in the nematode C. elegans. | ||
|
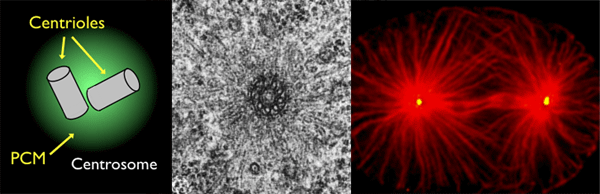
Microtubules are arranged at specific cellular sites called microtubule organizing centers (MTOCs). The vast majority of what is known about MTOCs comes from work on the centrosome, a non-membrane bound organelle composed of two centrioles surrounded by a cloud of pericentriolar material (PCM). The centrosome serves as an MTOC in dividing animal cells where microtubules are organized into radial arrays by proteins embedded within the PCM. | |
 |
Non-centrosomal MTOCsIn many types of differentiated cells such as neurons, myotubes, and epithelial cells, microtubules (red) are organized into non-radial arrays at non-centrosomal sites. These non-centrosomal MTOCs (ncMTOCs, blue) are a key feature of differentiated cells and vary by tissue type and organism. However, unlike the centrosome, far less is known about the proteins that comprise ncMTOCs and the mechanisms by which they are formed during development. Using novel genetic and proteomic tools in the model organism C. elegans (see below), we aim to understand these two main aspects of ncMTOC biology. |
|
 |
Centrosome InactivationMTOC activity at the centrosome is biphasic, cycling through assembly and disassembly during the cell cycle. Upon cell differentiation, the centrosome is often inactivated and MTOC function is reassigned to non-centrosomal sites to accommodate different cell functions. Although hyperactive centrosomal MTOC activity is a hallmark of some cancers and has been linked to invasive cell behavior, little is known about how the centrosome is inactivated as an MTOC either during mitotic exit or maintained in an inactive state in differentiated cells. We are using C. elegans as a model to understand these fundamental knowledge gaps. Our analysis of PCM proteins (e.g. SPD-2/CEP192 and SPD-5) in cycling embryonic C. elegans cells revealed that the centrosome is inactivated as an MTOC by a two-step process beginning with PCM 'dissolution', followed by mechanically controlled 'rupture'. We are now uncovering the control mechanisms and targets within the centrosome that underlie this two-step model in both cycling and differentiated cells. |
|
 |
Epithelial cell polarityThe ability of cells to polarize, or asymmetrically localize proteins, nucleic acids, lipids, and other macromolecules to specific cellular regions, underlies development and tissue homeostasis and goes awry in numerous human diseases. Epithelial cells line metazoan organs and are polarized along an apicobasal axis, which is critical for selective barrier and transporter functions, and for mechanical resiliency. Due to the importance of epithelial polarization to cell, organ, and organismal function, a wealth of studies have uncovered proteins required for this process, including the conserved apical polarity complex PAR3/PAR6/aPKC and the epithelial cell cadherin/catenin adhesion complex. Surprisingly, however, basic mechanistic features controlling epithelial polarization, especially in vivo (i.e. within organisms) are still unknown. In particular, how epithelial cells ‘break symmetry’ and designate a particular site that will direct the formation of a future apicobasal axis and how this information is used to target specific proteins to cellular surfaces are key knowledge gaps that we are using the C. elegans embryonic intestine to study. |
|
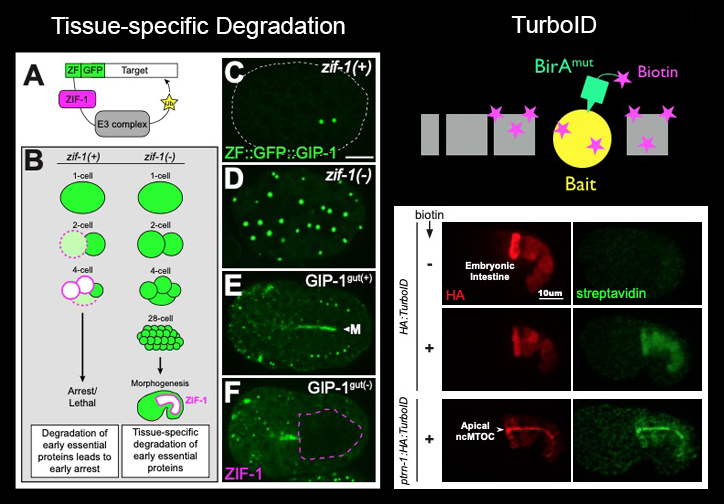 |
Genetic & Proteomic ToolsTissue-specific degradation: We have optimized a two-component system pioneered by the Nance Lab to deplete endogenous proteins from C. elegans tissues (left). By inserting a small ZF-degron tag into endogenous loci using CRISP/Cas9 and expressing the degrading protein ZIF-1 in a zif-1 mutant background, we can remove early essential proteins such as the γ-TuRC component GIP-1/GCP3 from differentiated cells.TurboID: In collaboration with Alice Ting's lab, we have developed the proximity labeling approach TurboID for use in C. elegans. TurboID catalyzes the addition of biotin to proteins in close proximity in just 10 minutes! In this example, TurboID labels proximity interactors of the bait protein PTRN-1/Patronin, a component of the apical ncMTOC in embryonic intestinal cells. |
|
 |
New HorizonsWe are using the amazing arsenal of tools now available in C. elegans to explore new directions in cell and developmental biology beyond what is listed above. These include cell size regualtion, epithelial connectivity, and mechanobiology. |
|