Sindy K.Y. Tang, Ph.D.
Stanford University

Engineering physical context to reveal biological function. Biological function is shaped not only by molecular components, but also by physical context, including geometry, mechanics, spatial organization, and history. We study how these physical factors carry biological information that governs function, repair, and adaptation. To do so, we develop tools that preserve or expose physical and spatial context, enabling functional, spatial, and mechanistic biological insights that are inaccessible with conventional workflows.
Our research spans foundational studies that define how physical context encodes biological behavior, to translational and synthetic efforts that exploit this context for measurement, prediction, and design. Current and recent projects include:

How geometry, mechanics, and history encode cellular state. Some single-celled organisms exhibit extraordinary biological "superpowers", such as the ability to survive catastrophic damage and rebuild themselves. We study how defined physical perturbations govern repair and experience-dependent adaptation. Using precise mechanical wounding, live-cell imaging and proteomic analysis in the giant unicellular organism Stentor coeruleus, we link physical context to molecular and functional signatures of cellular memory.
Cellular behavior is shaped not only by molecular composition, but also by physical context and history. We study how controlled physical perturbations, such as reproducible wounding and geometric constraints, influence subsequent repair, resilience, and adaptive responses in single cells.
Our work focuses on Stentor coeruleus, a giant unicellular organism capable of regenerating from fragments smaller than 1/27 of its original size. To precisely define physical context, we developed a microfluidic "guillotine" that enables reproducible control of wound geometry and damage conditions in living cells. This platform allows us to probe how prior physical experiences influence future responses, providing a rigorous framework to study cellular memory and experience-dependent adaptation without invoking cognition.
Building on this foundation, we are employing live-cell imaging and post-perturbation proteomics to capture how defined physical experiences reshape repair programs. We are also extending mechanical dissection and spatial analysis approaches to the scale of a single giant cell, enabling spatially resolved interrogation of how internal organization, damage, and repair are coordinated within one cell.
This work also informs our science outreach initiative, Nature's Tiny Marvels, which highlights the surprising "superpowers" of microscopic life as an entry point to understanding fundamental biology.
Relevant papers: Science 2017, PNAS 2017, BMC Biology 2021, Chem. Rev. 2022, SciRep 2024.
Funding: NSF
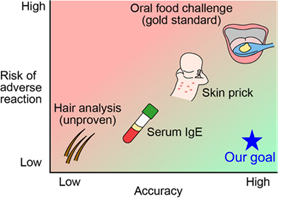
Preserving physical and temporal context in allergy diagnostics. Immune function is highly sensitive to physical and temporal context, yet much of this information is lost during conventional sample handling. We develop front-end engineering strategies that capture and stabilize immune function at the moment of blood collection. By locking in time-sensitive biological states and pairing these workflows with machine-learning-enabled analysis, we enable more physiologically meaningful measurements for food allergy diagnosis and monitoring.
Food allergy diagnosis is fundamentally a problem of measuring immune function, but standard workflows often degrade functional information before analysis. Our work focuses on preserving the physical and temporal context that governs basophil reactivity, enabling more accurate, safe, and clinically meaningful diagnostics.
We develop engineered sample-preparation strategies for basophil activation testing (BAT) that stabilize immune cells immediately after blood draw, preserving reactivity over extended handling and transport. We pair these workflows with machine-learning-based analysis to reduce subjectivity in gating and extract robust functional phenotypes.
Beyond conventional activation markers, we use label-free approaches and basophil migration to capture functional aspects of immune sensitivity and reaction severity. These complementary readouts expose immune phenotypes that are difficult to access with standard assays, enabling improved diagnosis and monitoring of allergic disease and immunotherapy response.
Relevant papers:
Biomicrofluidics 2020, Lab on a Chip 2022, Allergy 2023,
Advanced NanoBiomed 2024, JACI 2024,
LoC 2025, Sensors & Actuators B 2025

Mechanical dissection and spatial mapping of complex biological systems. In multicellular systems, biological function is encoded not only in molecular composition, but also in spatial organization that is often lost during tissue dissociation. We develop mechanical dissection and spatial registration tools to preserve and map spatial context, enabling functional and molecular analyses with retained spatial meaning, including applications in cancer organoids and spatial proteomics.
In tissues, spatial organization encodes critical information about local interactions, microenvironments, and history. Conventional tissue processing often destroys this information through homogenization or dissociation. We engineer mechanical dissection tools that preserve spatial context at the front end of biological analysis.
Our work includes the development of micro-dissection technologies that partition fresh tissues into spatially indexed microstructures for downstream applications such as patient-derived organoid culture, drug screening, and biobanking. By standardizing the physical dissection process, these tools reduce pre-analytical variability and improve reproducibility. In parallel, we are developing scalable platforms for spatially resolved, mass spectrometry-based proteomic analysis of tissues, enabling deep, unbiased mapping of protein distributions while retaining spatial indexing.
Beyond tissues, we extend these approaches to microbial communities and other heterogeneous or mechanically complex matrices that are difficult to section using laser microdissection, enabling spatial analysis in systems that are otherwise challenging to access.
Relevant papers: MINE 2024, EML 2025, BioRxiv 2025.
Funding: NCI
Can we better understand the rules of life by building a synthetic cell or tissue bottom up? In selected exploratory studies, we investigate whether defined physical structure and constraints are sufficient to generate biological function. By introducing engineered geometry and mechanics into minimal or synthetic systems, we probe how physical context alone can drive organization, robustness, and behavior.
These exploratory efforts test the sufficiency of physical context in biological organization by introducing defined mechanical and geometric constraints into minimal or synthetic systems. By isolating physical structure from biochemical complexity, we ask which aspects of architecture are necessary to support function.
Read more
This work complements our measurement-driven research by informing how physical context shapes biological behavior and by providing design principles for biohybrid systems and engineered living materials. This area is intentionally exploratory and evolves with new tools and insights, with potential applications in biohybrid systems for therapeutics and "living" materials.
Funding: NSF
Droplet microfluidics, in which micro-droplets serve as individual reactors, has enabled a range of high-throughput biochemical processes. Initial research on this technology demonstrated 1000-fold increase in throughput and up to 1 million-fold reduction in cost compared with state-of-the-art screening methods using microtiter plates. Our capability to form highly uniform drops at > 1kHz also enables us to generate complex fluids and soft matter bottom-up, and study how the microscopic structure of the material affects its bulk properties. Our research involves both fundamental and applied aspects:
Project: Flow physics, collective behavior & machine learning of croweded droplets in confinement
Although single drops have been studied extensively, the flow of concentrated emulsions—where droplet volume fraction exceeds 80%—is relatively unexplored in confined microfluidic systems. The many interacting interfaces in this regime give rise to complex behavior that cannot be predicted by dilute systems. Prior work on concentrated emulsions has focused on bulk rheology. The behavior of individual drops within the emulsion is unknown, but is critical as each drop carries a different reaction. We have developed experimental and analytical methods to elucidate the dynamics and fate of individual drops within a concentrated emulsion, and the identification of emergent behavior.
Relevant papers:
PRF 2022,
PNAS 2021,
Soft Matter 2019,
PNAS2016,
Soft Matter 2014.
Project: Interfacial mass transport: nanoparticles (NP) as droplet stabilizers to suppress crosstalk
Droplet-based biochemical assays rely on the reagents to remain isolated in individual droplets. We observed that the surfactant necessary to stabilize the drops against coalescence also mediates the inter-drop transport of molecules. Recognizing that solid particles can also stabilize droplet interfaces, we demonstrated the synthesis of partially fluorinated silica nanoparticles (F-SiO2 NP) as effective stabilizers for aqueous drops in fluorinated oils. Replacing surfactants with NP suppresses crosstalk. The high desorption energy of NP (>10^3 kT) renders NP adsorption at the droplet surface irreversible. NP do not form micelles. A key pathway for inter-drop molecular transport—association with surfactant reverse micelles—is thus eliminated.
Relevant papers:
ACS Appl. Mater. Interfaces 2014, Anal. Chem. 2015, RSC Adv. 2016.
Project: Using micro-droplets as biochemical reactors
Applications: Detection of antibiotic resistance, pathogens for diagnosis of tuberculosis, and using bacteria to convert methane into bioplastics.
Relevant papers:
Sensors & Actuators B 2018, Biomicrofluidics 2015, Bioresource Technology 2016.