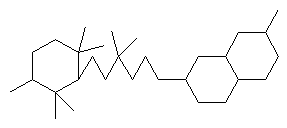Chemical Literature (Chem 184/284)
University of California at Santa Barbara
Lecture 15: Chemical Abstracts Registry File, Part 2: Structure Searching
Structure Searching on STN
- One of the most powerful features of the chemical substance files on STN is the
ability to search by chemical structure.
- A large number of STN files contain searchable chemical structures of various types.
- The REGISTRY, BEILSTEIN, GMELIN and Derwent Drug Files all contain records for
individual compounds.
| Input |
------> |
Output |
| Specific structure |
------> |
Single compound |
| Generic structure |
------> |
Set of compounds |
- The CASREACT, CHEMINFORMRX, “Derwent Journal of Synthetic Methods” and
CHEMREACT files all contain information on organic chemical reactions. The reactants
and products are structure searchable.
| Input |
------> |
Output |
| Specific or generic structures |
------> |
Set of reactions with desired features |
- The MARPAT and MARPATpreviews files contain the generic Markush structures appearing
in chemical patents in structure searchable form
| Input |
------> |
Output |
| Specific or generic structures |
------> |
Patents containing appropriate Markush structures |
How Structures are Stored
- In STN files, structures are stored as connection tables —
a list of each atom in the structure, which atoms each is linked to, and by what
kind of bond.
- Structures with stereochemistry have additional information about the spatial
arrangement of the bonds.
How Structures are Searched
- The Messenger software searches structure information in two steps: screening and
atom-by-atom match.
- Screening filters out likely matches by looking for certain common features.
- Atom-by-atom match then compares the whole of the connection table of the query
structure with that of the possible matches.
Building Query Structures
- Messenger has a whole set of commands for “drawing” chemical structures
within the system itself.
- STN Express is a specialized software package which includes structure
drawing software and the ability to upload these structures to the online system. For a
manual on building structures using STN Express, see
Structure Searching in the CAS Registry File
at http://www.cas.org/ACAD/casreg.pdf. Note: this is a large
PDF file requiring a current version of the Adobe Acrobat Reader for viewing.
Structure Building Commands — STRUCTURE
- The command STRUCTURE initiates the structure building process.
- The system responds by prompting for a structure to recall.
- You may respond with the name of a template, a Registry Number, a previously-built structure
L# or NONE.
- When in structure building mode, the arrow prompt is replaced by a colon.
GRAPH — Creating the Pieces
- The GRAPH command (abbreviated GRA) tells the system to create atoms or sets of atoms, as
either chains or rings. Note that, in general, you do not have to draw hydrogen atoms as
part of the structure—they are assumed to be present by the system.
- The default atom is carbon; the default bond is unspecified.
- When structure building with GRA commands, the system automatically assigns a number to
each node in the order constructed.
: gra c3
creates a three carbon chain, while
: gra r6
creates a six carbon ring. (i.e. the beginnings of cyclohexane or benzene)
: gra r66
creates two six membered rings fused along one side (i.e. the beginnings of naphthalene.)
- You can also attach chains to specific atoms:
: gra 2 c4
attaches a 4 carbon chain to atom 2
- You can create bonds between existing atoms:
: gra 1 2
DELETE — Removing Atoms and Bonds
- DELETE can be used to remove atoms or groups of atoms:
: del 1 5 8
- Or it can be used to remove bonds:
: del 1-2 7-9
NODE — Transmuting Elements
VARIABLE — Defining Your Own Generic Groups
BOND — Specifiying Bond Types
DISPLAY — Seeing What You've Built
- The DISPLAY command in structure building may be used at any step along the way to see
what the current structure looks like. It is frequently added to a structure building
command to save time.
: gra c3, nod 2 o, dis
- DIS SIA displays both the diagram and the attributes (see below) of the structure.
Attribute Commands
END — Going from Building to Searching
- When all your structure building is complete, the END command creates an L# for the structure
and returns you to the normal search mode.
- You must END a structure before you can search it.
Types of Structure Searches
- Messenger allows four types of structure search:
- EXACT: Looks for the compound exactly as drawn; only possible variations are
isotopic (and stereochemical if unspecified)
- FAMILY: Same as above, but will also pick up salts (of acids) or polymers (of
monomers).
- CSS: Stands for Closed Substructure Search This type of search will only allow
substitution where you have specfically allowed it, as with a CONNECT attribute
or the use of a variable or generic group.
- SSS: Stands for Substructure Search. Will allow any substitution at any atom
except as you have specifically restricted it.
Ranges of Searching
- You can also specify how much of the database you wish to search:
- SAMPLE: This is a fixed, randomly selected, 5% of the database. Always
search this before doing a substructure search to see if the search
will work. Sample searches are always free!
- FULL: Self-explanatory
- RANGE: You may specify a range of Registry Numbers to search; useful for update
searches or to continue searches which were to big to complete in one step.
Ranges of less than 100K RN’s are cheaper than a full search.
- SUBSET: Lets you use a previously created L# (by name, mol. formula, ring data,
structure or combinations) as the defined set to search on. Can be a very
powerful tool.
Structure Search Hints
- When doing a structure search, always use SEARCH, not S. This way, the system will
prompt you for type of search and range of search.
- SAMPLE searches aren’t necessary for EXACT or FAMILY searches, but are
strongly recommended for substructure searches.
- If a structure is unsearchable (exceeds system limits), consider whether you can create
a suitable subset with name fragments or molecular formula or ring information which
would bring a subset search within system limits. Alternatively, modify the structure
to make it more specific. Note that changing HCO or CON attributes does not affect the
search at the screening level, so these limitations do not generally keep a search within
system limits.
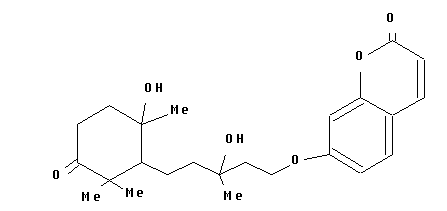
Structure Building Example: Feropolone
- First, build the rings:
:gra r6
:gra r66, dis
- Then, build the chain connecting the two:
:gra 1 c6
:gra 22 7
- Now, build the side chains:
:gra 2 c1, 2 c1, 3 c1, 5 c1, 5 c1, 11 c1, 19 c1, 19 c1, dis
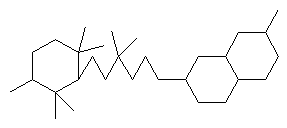
- Then, use the NOD command to change atoms as necessary:
:nod 10 22 25 28 o, 23 24 29 me, 27 30 oh, dis
- Now apply the BON command:
:bon all se, dis
:bon 3-25 11-28 12-13 de, dis
:bon 7-8 8-9 9-14 14-15 15-16 16-7 n; dis
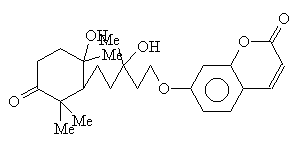
- Then apply attribute commands as necessary.
- When the structure is completed, use the END command to complete the structure
and return to the regular search mode.
- You may display completed structures online with “display query L#”
- Search the query with “search L# [search type] [search range]”
=> search L1 exact full
=> search L1 sss sample
EXAMPLE:
=> search l3
ENTER TYPE OF SEARCH (SSS), CSS, FAMILY, OR EXACT:sss
ENTER SCOPE OF SEARCH (SAMPLE), FULL, RANGE, OR SUBSET:full
FULL SEARCH INITIATED 23:13:37
FULL SCREEN SEARCH COMPLETED - 71 TO ITERATE
100.0% PROCESSED 71 ITERATIONS 1 ANSWERS
SEARCH TIME: 00.00.02
This page created by
Chuck Huber (huber@library.ucsb.edu).