
 |
 |
Helix-HIV > Helix Vectors and Maps
Helix-HIV lentivirus vectors
Background
The use of retroviral vectors for delivery of genes has been well established in the last few years. Their properties make them very attractive for use in gene therapy. They can accommodate long sequences, the products of which will be stably expressed due to integration into the cell chromosome, and seem to be non-immunogenic due to the lack of viral coding sequences transfer.
Moreover, vectors based on the lentiviruses subclass of Retroviridae have the special characteristic of being able to transduce non-dividing cells. But bearing in mind that the most characterized member of the lentiviruses that naturally infects human cells is HIV-1, there remain safety concerns. There are several means to overcome these concerns.
Non-RCR vectors
The production of viral particles in a producing cell line that will be able to infect target cells, but won’t be able to form a replication-competent recombinant (RCR) and subsequently infect other target cells is a feature behind the development of HIV-1 based vectors.
When HIV-1 based vectors are used, the goal has been to diminish the probability of a recombination event that will give rise to full-length replicative viral DNA. This has been accomplished by separating the structural and delivery constructs onto three different plasmids. The regulatory/accessory genes of HIV-1, specifically Nef, Vpr, Vpu, Vif and Tat have been shown to be largely dispensible for gene delivery to most cell types. The relevance of the need for the different proteins of HIV-1, particularly for gene therapy, namely infection efficiency of the target cell and expression level of the transgene, is beyond the scope of this short discussion. Gag- Pol and Env have been separated into two independent plasmids. These, and other safety features are fully discussed in the references listed below.
Sin vectors
Although the recombination event possible between three independent plasmids is extremely small, ways to decrease even further this probability when dealing with HIV-1 based vectors have been undertaken in the plasmids presented at this site. This is the basis of the development of self-inactivating vectors or SIN vectors.
Basically, most of the U3 region of the 3’LTR is deleted. This site harbors the major transcriptional functions of the HIV genome. During the process of reverse transcription, the 3’LTR is copied to the 5’LTR. By deleting non-replicative portions of the 3'LTR, the genomic viral DNA is inserted into the target genome as a promoter-less sequence. The lack of active viral promoter avoids both the possible transcription of the viral sequence and detrimental effects on eukaryotic gene expression.
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998. 72(12):9873-80.
- Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998. 72(10):8150-7.
HIV-1 genome:
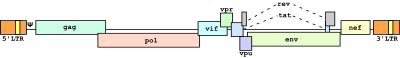
Structure of a non-RCR-HIV-1 based vector

Construction of the sin-HIV-1 based vector

- The picture shows all the proteins of HIV-1 are driven by the CMV promoter. For the actual production of virus, most of the proteins are deleted, as in the second and third generation of HIV-1 based vectors.
- Due to the extremely low efficiency of pseudotyping HIV-1 with HIV-1 envelope in trans, envelopes of other viruses have been employed efficiently, such us GalV, VSV-G, and MLV.
- Different versions of the delivery vectors (see Helix vectors).
Helix vectors
Some of the vectors exist in different versions. The vectors described here contain the psi sequence including the 5’ of gag. A short version of psi also exist. Although the CMV promoter is the one depicted here, some vectors have been made with the RSV promoter instead. The marker insert might be other than GFP, and some plasmids exist with the marker in both orientations.
These plasmids are provided as examples only--THEY ARE NOT CURRENTLY AVAILABLE. When we make the vectors available to the research community they will be a set of tested and verified constructs.
-Basic pHs vector
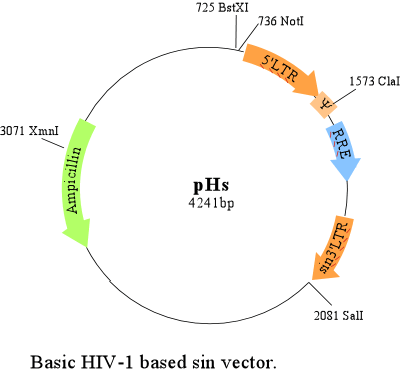
Download sequence of above
-pHs-CG vector
This is the basic vector with the eGFP gene driven by the CMV promoter

Download sequence of above
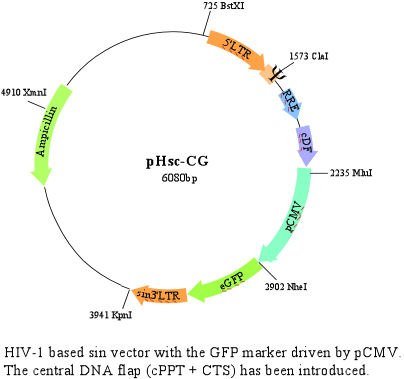
-pHsc-CG vector
This is basically the pH-CG vector with a cDNA PPT flap.
The lentiviruses, HIV-1 among them, are unique in their ability to infect nondividing cells. This feature is based on a nuclear import pathway that enables the viral DNA to cross the nuclear membrane of the host cell. In the process of HIV-1 reverse transcription, a plus strand overlap; the central DNA flap, is created following a strand displacement in the center of the genome. This is the result of synthesis initiation from the central polypurine tract; cPPT, and termination of plus strand synthesis by the central termination sequence, CTS. The DNA flap seems to act in cis to enable HIV-1 DNA nuclear import. Introduction of this element into the plasmid is thought to avoid or at least decrease the accumulation of DNA in the cytoplasm. (Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000. 101:173-85.)
sequence (INTRODUCE Seq 6)
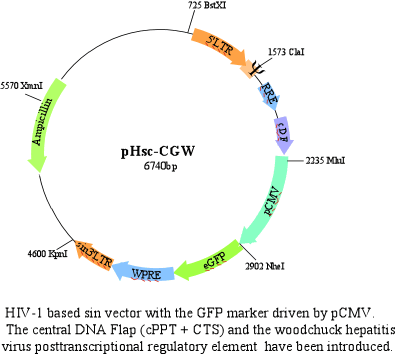
-pHsc-CGW vector
This is the pHc-CG vector with the WPRE.
The WPRE seems to increase the expression of the marker gene when introduced in its 3' untranslated region. This element is believed to exert some posttranslational regulation.
-Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999. 73(4):2886-92.
sequence (INTRODUCE Seq 7)
Related links:
Helix MTAs
Home Page |
Interests | Members
|
MTA Forms | Plasmid
Maps | Retroviral Systems
| Genetic Screens | Library
Systems | Protocols
| Tutorials |
Publications | Contact
| Virus Chat