Research Discussions:
The following log contains entries starting several months prior to the first day of class, involving colleagues at Brown, Google and Stanford, invited speakers, collaborators, and technical consultants. Each entry contains a mix of technical notes, references and short tutorials on background topics that students may find useful during the course. Entries after the start of class include notes on class discussions, technical supplements and additional references. The entries are listed in reverse chronological order with a bibliography and footnotes at the end.
June 15, 2013
We spent the last three months studying how to get detailed information out of the brain at sufficient quantity and quality that would make it interesting, even necessary to apply industrial-scale computing infrastructure to analyze and extract value from such data. If we were thinking about launching startup, the next step would be to formulate a business plan to define and motivate a specific project and the staffing required to carry it out. Concretely, we might start by taking a close look at the investment opportunities identified in the report we produced in CS379C this quarter.
As an exercise, I will tell you how I would tackle the problem of formulating a business plan for launching a project at Google. I’m using the term “business plan” rather broadly here to include prospects for scientific research; the “business” aspect of the plan is that the project has to make sense for Google to invest effort. Specifically, it has to be something that has significant societal or scientific merit and only Google could carry it off by leveraging its infrastructure and expertise, or it offers new opportunities for growth in existing product areas or developing new revenue streams that make sense for Google financially and technologically.
In the following, I’ll elaborate on an idea we talked about in class and that I believe has promise by offering (a) a new approach to computational neuroscience that leverages what Google does best — data-driven analytics and scalable computing, (b) opportunities to make significant advances in the knowledge and methodology of neuroscience, and (c) the possibility of discovering new algorithms for machine learning and pattern recognition and new approaches for building efficient computing hardware.
I maintain, based on investigations over the last nine-months and discussions with some of the best systems neuroscientists in the world that we will soon — within at most a couple of years — have the ability to record from awake, behaving mammals — mouse models are just fine — data that can be used to recover spikes at unprecedented spatial and temporal scale.
I’m currently focusing on two technologies that we talked about in class. The first technology involves advances in 3-D microarrays for electrophysiology that leverage CMOS fabrication to achieve 10 micron resolution in z, 100 micron resolution in x and y, and millisecond temporal resolution [51]. New optogenetic devices may supersede this technology in a few years with more precise tools for intervention and new options for recording [176], but for now we can depend on a relatively mature technology accelerated by Moore’s law to sustain us for the next couple of years.
The second technology involves the use of miniaturized fluorescence microscopes that offer ~0.5 mm2 field of view, and ~2.5 micron lateral resolution according to Ghosh et al [59]. The fluorescence imaging technologies have lower temporal resolution and more difficulty in recovering accurate spike timing [160], but offer better options for shifting the field of view, i.e., could be done mechanically or optically versus removing and then inserting the probe in a new, possibly overlapping location as would be required in the case of an implanted 3-D probe, and less tissue damage and potential for infection limiting the effective duration of chronic implants.
Collecting data would precede as follows: First, identify a 3-D volume of tissue in the experimental animal’s brain which we’ll refer to as the target circuit and position the recording device so that it spans a somewhat larger volume completely enclosing the target circuit. Identify the “inputs” and “outputs” of the target circuit, which for the purpose of this discussion we assume correspond to, respectively, axonal and dendritic processes intersecting the boundaries of the target. Collect any additional information you may need for subsequent analysis, e.g., we might want to identify capillaries to use as landmarks if there is a plan to subsequently sacrifice the animal and prepare the neural tissue for microscopy in order to collect static connectomic and proteomic data.
Record as densely as is feasible and especially at the boundaries of the target tissue while the experimental animal is exposed to test stimuli aligned with the neural recordings for subsequent analysis. Note that it isn’t necessary to record from specific cells or locations on cells, e.g., a uniformly spaced grid of recording locations, while implausible, would work fine for our purposes. The above-mentioned recording technologies should make it possible to collect extensive recordings of each animal, and running many animals in parallel should be possible using modern tools for automating experimental protocols [38, 175]. Finally, if applicable, prepare the tissue for staining and scanning to obtain additional provenance relating to structure (connectomics and cell-body segmentation) and function (proteomics and cell-type localization).
Now we want to learn a function that models the input-output behavior of the target tissue. Specifically, we are interested in predicting outputs from inputs in the held-out test data. I don’t pretend this is going to be easy. In the simplest approach we would ignore all the data except on the target boundaries. Knowing the types of cells associated with the inputs and outputs might help by constraining behavior within cell-type classes. In a more complicated approach, we might be able to recover the connectome, use it to bias the model to adhere to connectomically-derived topological constraints and treat the sampled locations other than those on the boundary as partially-observable hidden variables. As I said, this isn’t likely to be easy, but rather than trying to use theories about how we think neurons work to bias model selection, I’m interested in seeing what we can accomplish by fitting rich non-parametric models such as the new crop of multi-layer neural networks [91, 94].
Given how hard this problem to likely to turn out in practice, I think it wise to start out on training wheels. I suggest we begin by developing high-fidelity simulations for in silico experiments using technology such as MCell which is able to incorporate a great deal of what we know about cellular processes into its simulations [143, 37]. Unlike the attempts at simulating a single cortical column conducted by the EPFL / IBM Blue Brain collaboration, we envision starting with an accurate model of cytoarchitecture obtained using electron microscopy coupled with proteomic signatures synapses obtained from array tomography. In any case, we would want to start with much simpler target circuits than a cortical column consisting of 60,000 cells.
Once we have a reasonably accurate simulation model for target circuit, the next step would be to “instrument” the model to simulate the technologies we plan to use in our in vivo experiments. Since our models are at the molecular level it should be possible in principle simulate electrophysiology and calcium imaging. Given the resulting simulated version of our experimental paradigm, we would proceed to collect data controlling for the size and complexity of the target circuits as well as our ability to accurately record from and collect supplementary structural and functional meta data. A first step for a startup would be to build or adapt the requisite tools and create the models necessary to perform such in silico experiments and demonstrate some initial successes in recovering function from simulated neural tissue.
P.S. July 16, 2013: Most of the students in this year’s class had engineering backgrounds and knew basic electromagnetic and electrical-circuit theory, but I did have some questions about electrophysiology and biological neural models from people who ran across these pages on the web. I told them enough to answer their immediate questions and pointed them to some relevant textbooks and Wikipedia pages to continue their education. Below is a mashup of my responses included here as a starting point for other visitors to these pages — most of the content is in the footnote. If this is too much technical detail, you might consider the electric-hydraulic analogy which for all its shortcomings often helps to make simple circuits more comprehensible and intuitive.
The primary tools of the electrophysiologist include the voltage clamp which is used to hold the voltage across the cell membrane constant so as to measure how much current crosses the membrane at any given voltage, the current clamp in which the membrane potential is allowed to vary and a current is injected into the cell to study how the cell responds, and the patch clamp which is a refinement of the voltage clamp that allows the electrophysiologist to record the currents of single ion channels. To understand the technical details of these methods, you will need a basic understanding of elementary electromagnetic theory1
June 9, 2013
I asked Justin Kinney, a postdoc in Ed Boyden’s lab, about using their new probes for recording from mouse cortical columns. Here’s what he had to say followed by my posting to CS379C and Mainak’s reply:
JK: Our custom silicon probes are planar arrays of 64 to 128 gold pads per shank (100 microns wide by 15 microns thick and 1-2 mm in length). The pads are 10 microns in a side with a 15 micron pitch, with the possibility of shrinking both dimensions. We are experimenting with different ways to maintain a low electrical impedance of the pad, e.g., 1 Mohm, even as we shrink the size of the pads. To keep tissue damage at reasonably low levels, we must space the shanks pretty far apart, e.g, 100s of microns. So the question of brain coverage — can we record from every neuron in a volume — becomes a question of the “field of view” of an individual shank. How large is the volume of brain tissue around a probe for which the activity of every neuron is accurately recorded? We do not know the answer to this question yet. We have experiments planned which should give us some clues.TLD: So, it seems that resolution in z — along the shank — is pretty good (15 microns) but in x and y it’s pretty poor (100’s of microns). The 3-D optogenetic device described in Zorzos et al [176] looks to have similar constraints though the probes are somewhat thicker (65 microns) presumably to accommodate the light guides. I would think calcium imaging using the Schnitzer lab’s miniaturized fluorescence microscopes would be a better option in trying to instrument a cortical column, ~0.5 mm2 field of view, and ~2.5 micron lateral resolution according to the Ghosh et al 2011 paper. Lower temporal resolution I expect but perhaps less problematic in terms of shifting the field of view, i.e., could be done mechanically or optically versus removing and then inserting the probe in a new location as would be required in the case of the device that Justin describes.
What is the temporal resolution and is it dominated by the microscope hardware, GECI characteristics or the computations involved in spike-sorting and sundry other signal processing chores? Lateral resolution could be improved significantly with better CMOS sensors according to Ghosh et al [59]. What do you think?
MC: I think the characteristics of the transient Calcium response (i.e. kinetics of the GECIs) are more important than microscopic hardware or the algorithms employed. All localizing (or spike sorting) algorithms I have seen for spike train recovery use some sort of a deconvolution step which essentially amounts to undoing the spread due to the non-ideal kinetics (see here for representative response curve). This is regardless of the sophistication of the microscope or algorithms.
Improving the microscopic hardware may give us better fields of view (thereby helping us scale up the number of neurons), algorithms may give us better ability to distinguish cells based on patterns (spatial filters for image segmentation), but when it comes down to recovering precise spike timings of a population of neurons from filtered waveforms, I think that the time spread due to calcium kinetics impose critical limitations.
As an aside, there is a body of work called finite-rate-of-innovation signal recovery (e.g. see here) which can help us recover the amplitudes and time shifts of waveforms like ∑iN ai exp ( - (t - ti)). This is a constrained model, but one thing it tells us is that if the number N of independent neurons is known, one can recover the spike timings precisely (thereby implying that GECI kinetics are not a limitation anymore for temporal resolution). However I am not sure how robust this would be to noise or model imperfections.
JK: Globally, yes, the x and y resolution, defined as the distance between shanks of a multi-shank probe, is 100s of microns (as we imagine it now to keep tissue damage low). However, on a single shank, we tighly-pack pads in multiple columns, likely 2 to 4. Such a probe will sample the brain in globally-sparse, locally-dense way. I attached a photo taken through a microscope of a two-column shank.
As you point out, calcium imaging typically has larger fields of view than electrophysiology probes. On the other hand, calcium-indicator dyes generally have poor temporal response (even GCaMP6) and low-pass filter the neural activity. We are working on an experiment to perform simultaneous two-photon calcium imaging and ephys recordings (with our custom probes) to better understand the limitations of each technology.
June 7, 2013
Here are some follow-up notes from Thursday’s class addressing questions that were raised in discussions relating to Daniel’s and Mainak’s projects. The first topic relates to recycling and garbage collection in the brain:
Inside the cell, organelles called lysosomes are responsible for cleaning up the debris — digesting macromolecules — left over from cellular processes. For example, receptor proteins from the cell surface are recycled in a process called endocytosis and invader microbes are delivered to the lysosomes for digestion in a process called autophagy. Lysosomes contain hydrolase enzymes that break down waste products and cellular debris by cleaving the chemical bonds in the corresponding molecules through the addition of water molecules.
In the extracellular matrix, macrophages take care of garbage collection, by ingesting damaged and senile cells, dead bacteria, and other particles tagged for recycling. Macrophages tend to be specialized to specific tissues. In the central nervous system, a type of glial cell called microglia are the resident macrophage. In moving through extracellular fluid in the brain, “if the microglial cell finds any foreign material, damaged cells, apoptotic cells, neural tangles, DNA fragments, or plaques it will activate and phagocytose the material or cell. In this manner, microglial cells also act as ‘housekeepers’ cleaning up random cellular debris” (source).
Following our class discussion about toxicity and metabolic challenges induced by adding cellular machinery for recording, I got interested in reviewing a lecture by Robert Sapolsky entitled “Stress, Neurodegeneration and Individual Differences” from his “Stress and Your Body” series [134]. Sapolsky notes — around 33:30 minutes into the video — that “over and over again in neurological insults, all hell breaks loose, you can still carry on business pretty well, you just can’t afford to clean up after yourself”, by which he means intracellular housecleaning.
Relating housecleaning to stress, the point is that stress-induced levels of glucocorticoids damage the hippocampus by altering the ability of its cells to take up energy and then causing the cells to do more work generating waste products in the process, thereby leaving the cell depleted of energy and littered with waste that would require even more energy to recycle or expel from the cell if the cell could afford it. The cell is left exhausted and at the mercy of apoptotic and autophagic mechanisms responsible for induced cell death.
Sapolsky begins by describing the cascade of physiological events precipitated by a stress-inducing stimuli. The first steps involve the hypothalamus which contains a number of small nuclei that serve to link the nervous system to the endocrine system via the pituitary gland. In the classic neuroendocrine cascade, the hypothalamus releases CRF and related peptides which stimulate the pituitary to releases ACTH which stimulates the adrenal glands to release glucocorticoids — see here for a discussion of the hypothalamic pituitary adrenal axis.
Glucocorticoids inhibit glucose uptake and glucose storage in tissues throughout the body except for exercising muscle. The mechanism — described 27:30 minutes into the video — works as follows: Glucocorticoids bind to their receptors on the cell surface, translocate to the nucleus where they induce the production of a protein called sequestrin that sequesters glucose receptors off of the cell surface in a process akin to endocytosis, sticking them into “intracellular mothballs”, and resulting in a cell that doesn’t take up as much glucose.
Sapolsky’s lab determined that — in the case of neurons — the reduction in glucose uptake is on the order of 20% and, in the case of sustained chronic stress, this level of reduction inevitably leads to cell death. The reason I mention this bit of esoteric knowledge here, apart from the fact that the related science is interesting, is because this sort of interaction is quite common in neuropathologies and underscores the dangers of stressing neurons with their tightly-controlled metabolic budgets.
The second topic concerns the problem of instrumenting cells in specific neural circuits for the purpose of targeted recording and relates to some issues raised by Mainak:
The cortical minicolumn is a vertical column of cortical tissue consisting of up to several hundred neurons. The minicolumn and related macrocolumn — also called hypercolumn or cortical module — has been the subject of several interesting and often controversial hypotheses about cortical computations the best known of which is generally credited to Vernon Mountcastle [116, 115, 114, 113] — the Wikipedia article is technically wanting but the Buxhoeveden and Casanova review article [28] is reasonably thorough.
We briefly discussed the minicolumn as a target circuit that would exercise near- to medium-term recording technologies, and be small enough that it might be possible to infer something interesting about its function from detailed recordings. The challenge we debated was how to record from exactly one minicolumn in an awake, behaving model such as a mouse. A minicolumn is on the order of 28-40 μm and most 2-D microelectrode arrays (MEA) are on the order 5-10 mm and the latest 3-D optogenetic array from Boyden’s lab is on the order of a couple of millimeters with about 150 μm resolution along one of the x or y axis and perhaps a tenth of that along the z axis and the other x or y axis [176].
There is a collaboration of physicists from Drexel University and neuroscientists from the Boston University Medical Campus studying the spatial organization of neurons obtained from thin-sliced brain tissue sections of rhesus monkey brains to better understand the organization of macrocolumns and minicolumns and the connection between loss of such organization and cognitive declines seen in normal aging [39]:
It has [...] been noted that different cortical regions display a “vertical” organization of neurons grouped into columnar arrangements that take two forms: macrocolumns, approximately 0.4-0.5 mm in diameter [114], and [...] minicolumns approximately 30 microns in diameter [75].Macrocolumns were first identified functionally by Mountcastle [...], who described groups of neurons in somatosensory cortex that respond to light touch alternating with laterally adjacent groups that respond to joint and/or muscle stimulation. These groups form a mosaic with a periodicity of about 0.5 mm. Similarly, Hubel and Wiesel [...] using both monkeys and cats discovered alternating macrocolumns of neurons in the visual cortex that respond preferentially to the right or to the left eye. These “ocular dominance columns” have a spacing of about 0.4 mm. In addition, they discovered within the ocular dominance columns smaller micro- or minicolumns of neurons that respond preferentially to lines in a particular orientation.
Once these physiological minicolumns were recognized, it was noted that vertically organized columns of this approximate size are visible in many cortical areas under low magnification and are composed of perhaps 100 neurons stretching from layer V through layer II. To prove that the [...] morphologically defined minicolumns [...] are identical to the physiologically defined minicolumn would require directly measuring the response of a majority of the neurons in a single histologically identified microcolumn, but this has yet to be done. (source)
Probably worth thinking about whether any of the existing MEA technology would be of use in recording from a mouse minicolumn, or whether alternative technology like the Schnitzer lab’s micro-miniature (~2.4 cm3) fluorescence microscope would make more sense given its ~0.5 mm2 imaging area and maximum resolution of around 2.5 microns [59]. As for a method of selecting exactly the cells in a cortical minicolumn, apart from just recording densely from a region large enough to encompass a minicolumn, subsequently sacrificing the animal and then scanning the tissue at a high-enough resolution to identify the necessary anatomical details, I can’t think of anything simple enough to be practical.
June 5, 2013
Andre asked a professor from University of Texas Austin about some of the RF issues and his answers might help those of you working on related technologies to organize your thoughts and identify important dimensions of the problem.
AE: I’m taking a class in neuroscience in which we’re employing nanotechnology ideas to solve the problem of getting data out of the brain. Imagine we have a small optical-frequency RFID chip in each brain neuron, constantly sending out information about whether the neuron is firing or not, in real time. There are 1011 or 100 billion neurons, that is, 1011 RFIDs sending information optically to some receiver outside the skull.Do you happen to know of a sophisticated scheme which might allow us to deal with the issue of reading in 1011 transmitters sending information between ~214 THz and 460 THz (the biological ‘transparency’ window of 650-1400 nm wavelengths). I can envision frequency division multiplexing with (460 THz – 214 THz) / 1011 neurons = 2460 Hz per neuron, but I may be neglecting other issues, such as SNR or the ability to transmit in such a narrow window at such high frequencies.
BE: Large-scale problem. Fun exercise to think through despite the many current insurmountable barriers for such a system.
Scenario #1: Sensors are transmitting all of the time: For frequency division multiplexing, you had mentioned a transmission bandwidth of 2460 Hz per sensor. Assuming sinusoidal amplitude modulation, the maximum baseband message frequency would be 1230 Hz. If this were digital communications, then the sampling rate would be 2460 Hz at the sensor of the neural activity.
Scenario #2 — On-Off Keying: To save energy, each sensor could use on-off keying. Only transmit something when the neuron has a spike. It saves on transmit power and reduces interference with other transmitters. You could still use frequency division multiplexing as before.
Scenario #3 — Wishful Thinking: Since we’re assuming the design, fabrication and implantation of 1011 sensors, we could assume the design and fabrication of 1011 receivers. Each would be tuned to one implanted sensor. This would help workaround the problem of a single receiver trying to sort out 1011 transmissions, which is not practical.
Scenario #4 — Frequency Allocation: It would be helpful if the transmission frequency band for a particular sensor is not close to the transmission bands of the sensors closest to it. There is always some leakage into other bands. For on-off keying, I recall that the leakage strength reduces versus frequency away from the sensor’s transmission band.
June 3, 2013
The audio for our discussion with Mark Schnitzer is now linked off the course calendar page. Thanks to Nobie and Anjali for recalling Mark’s references to Stanford scientists doing related work:
Ada Poon’s group with respect to RF techniques in tissues,
Bianxiao Cui’s group with respect to cultured neuron interfaces,
Nick Melosh’s group with respect to cultured neuron interface, and
Krishna Shenoy’s group with respect to neural prostheses in primates.
The reference to Ada Poon’s lab led me to several papers incuding this review of methods for wireless powering and communication in implantable biomedical devices [169].
We talked briefly at the end of class about the possibility of giving up on “wireless” and instead trying to figure out a way of installing a network of nanoscale wires and routers in the extracellular matrix. Such a network might even be piggybacked on the existing network of fibrous proteins that serve as structural support for cells, in the same way that the cable companies piggyback on existing phone and power distribution infrastructure. There was even a suggestion of using kinesin-like “linemen” or “wire stringers” that walk along structural supports in the extracellular matrix to wire the brain and install the fiber-optic network.
Perhaps it’s a crazy idea or, as someone pointed out, perhaps someone will figure out how to do this and, in the future, it will just be “obvious” to everyone. I did a cursory search and, in addition to some related ideas from science fiction stories, I found this Cold Spring Harbor Laboratory announcement of the 2013 Wiring the Brain Meeting scheduled for July 18 through July 22. It’s about how the brain’s neural network is wired during development; not exactly what I was looking for, but a timely reminder that nature might have some tricks worth exploiting. I also found this 2009 review paper [90] on nanomaterials for neural networks that has interesting bits about toxicity and nanowires.
Anjali and I talked after class about differences in the terminology surrounding ultrasound, specifically the acronyms for therapeutic ultrasound technologies — FUS, HIFU, USgFUS and MRgFUS and low-intensity US for stimulation — versus the diagnostic/imaging ultrasound technologies. I asked her to send me the names of a relevant contacts at Stanford and she supplied: Dr. Kim Butts-Pauly, Dr. Pejman Ghanouni, Dr. Urvi Vyas, and Dr. Juan Plata-Camargo.
We also discussed the ambiguity concerning the “imaging” category used in the paper, and Anjali suggested that I was “using imaging as a term encapsulating any externally recording, computing, and powered device rather than only the modalities that result in images” which I thought was a good characterization of my intent2.
May 31, 2013
It is no longer necessary to depend solely on what we discover in the natural world when searching for biomolecules for engineering purposes. Methods from synthetic biology like rational protein design and directed evolution [21] have demonstrated their effectiveness in synthesizing optimized calcium indicators for neural imaging [73] starting from natural molecules. Biology provides one dimension of exponential scaling and additional performance can be had by applying high-throughput screening methods and automating the process of cloning and testing candidates solutions. There are also opportunities for accelerated returns from improved algorithms for simulating protein dynamics to implement fast, accurate, energy function used to distinguish optimal molecules from similar suboptimal ones. Advances in this area undermine arguments that quantum-dots and related engineered nanoscale components are crucial to progress because these technologies can be precisely tuned to our purposes. Especially when weighed against the challenges of overcoming the toxicity and high-energy cost of these otherwise desirable industrial technologies.
Following Wednesday’s discussion, I’m a little concerned about how the analysis of the readout problem is progressing, i.e., the readout solutions that might be leveraged by advances in nanotechnology. In particular, I want you to be convinced by Yael’s and Chris Uhlik’s arguments that an RF approach doesn’t scale — in particular, see if you agree Chris’s back-of-the-envelope analysis as it seems to be more completely worked out than Yael’s analysis. Make sure you’re OK with their assumptions. If you agree with them, then it’s fine to say so and summarize their analysis and credit them in your report, e.g., use a citation like “Uhlik, Chris. Personal Communication. 2013”.
When and if you’re convinced, apply the same standards that you used in evaluating RF solutions in evaluating possible optical solutions. It’s not as though we can’t imagine some number of small RF transmitters implanted deep within the brain or strategically located inside of the dura but on the surface of the brain. These transmitters could number in the thousands or millions instead of billions and serve relatively large portions of the brain with additional local distribution via minimally-invasive optics of one sort or another — diffusion might even work if the distances are small. Broadcasting in the RF, these transmitters would have no problem with scattering and signal penetration depth. The problems with RF have more to do with bandwidth, practical antenna size for nanoscale devices and limited frequency spectrum available for simultaneous broadcast.
In the case OPIDs with LCD-mediated speckle patterns, even if you assume you could build a device that worked, i.e., could flash the LCD and some external imaging technology (perhaps inside the skull but outside the brain) could analyze the resulting light pattern and recover the transmitted signal, there is still the question of whether this would work with a billion such devices all trying to transmit at 1KHz. I can imagine how a small number of simultaneous transmissions might be resolved, but the computations required to handle a billion simultaneous transmissions boggles the mind — my mind at least, maybe you have a clever solution I haven’t thought of.
Make sure you’ve read Uhlik’s analysis and whatever Yael has in his slides and Adam’s notes, and then I’d like you to sketch your best solution and circulate it. Do it soon so we can all help to vet, debug, refine it, and bug our outside consultants if need be. Don’t be shy; just get something out there for everyone to think about. This is at the heart of scaling problem for readout and I’m not sure we have even one half-way decent solution on the table at this point.
By the way, to respond to Andre’s question, I have confidence that nanotechnology will deliver solutions in the long term. It isn’t a question of whether, only when. Funding will probably figure prominently in the acceleration of the field and right now biology-based solutions to biological problems have the advantage. Very successful biotech for photosynthesis-based solar power and extraordinarily efficient biofuel reactors are already eating into the nanotech venture capital pie and that could slow progress in the field. But there is room for growth and smart investors will understand the leverage inherent in harnessing extant chip design and manufacturing technologies to accelerate development:
AE: What role do you see venture capital playing in the near-term of this [neural recording] nanotechnology problem? It seems to me that things are largely still on the research end. Even when I look at the 2-5 year time frame, I can only see ventures such as non-toxic organic quantum dot mass production, assuming it is worked out.TLD: I’m thinking of a two-prong approach: (1) non-biological applications in communications and computing where biocompatibility isn’t an issue and current technologies are up against fundamental limitations, and (2) biological applications in which the ability to design nanosensors with precisely-controllable characteristics is important.
Regarding (1) think in terms of QD lasers, photonics, NFC, and more-exotic-entangled-photon technologies for on-chip communication — 2-D and 3-D chips equipped with energy-efficient, high-speed buses that allow for dense core layouts communicating using arbitrary, even programmable topologies.
Regarding (2) there is plenty of room for QD alternatives to natural chromophores in immunoflorescence imaging, voltage-sensing recording as discussed in Schnitzer’s recent QD paper in ACS Nano [103], and new contrast agents for MRI. The ability to precisely control QD properties will fuel the search for better methods of achieving biocompatibility and attract VC funding.
May 29, 2013
We discussed the format for the final projects so they would serve both for the individual student contributions to the jointly-authored technical report and also final-project document:
Three pages allotted for the jointly-authored technical report excluding bibliography:
For each technology deemed of sufficient merit for inclusion in the technical report or requiring discussion to offset unwarranted enthusiasm indicated in published reviews provide:
a summary description of the technology,
the rationale for inclusion in the report, and
a summary of the back-of-the-envelope analysis that led this rationale.
Additional pages as needed to cover the assigned topic for the final project:
For each technology considered of interest, whether or not it is included in the three pages for the technical report, provide:
a detailed description of the technology,
an extended rationale, and
the full back-of-the-envelope analysis with justification for any simplifications and order-of-magnitude estimates.
The latest version of the technical report includes drafts of the sections on “imaging”, “probing”, “sequencing” and new subsection on investment opportunities. Still left to go are the “automating” and “shrinking” sections, the latter of which we outlined in class identifying several areas requiring careful treating in the final version of the technical report. In particular:
Make clear the practical disadvantages of relying on diffusion as a mechanism for efficient information transfer in the cell contrary to its description as a solution in published descriptions of nanoscale communication networks.
Work out the issues concerning GECIs versus semiconductor-based quantum dots and related inorganic, compatibility-challenged non-biological solutions. This came up when it was noted that GECI technology and the GCaMP family in particular are improving at a rapid pace and the old arguments that natural biological agents are inferior to semiconductor-based quantum dots are being over turned as our ability to engineer better GECIs by accelerated natural selection matures [73].
What I was calling “accelerated natural selection” and “controlled evolution” in class is better known as protein engineering in the literature and generally qualified as pertaining to one of two related strategies: “rational protein design” and “directed evolution”. This was the method discussed in the paper from Janelia Farm on the optimizations that led to GCaMP5 [73].
Noting that any of the nanotechnologies we discussed would require some method “installation” and that suggesting we encourage the development of “designer babies” with pre-installed recording and reporting technologies would be awkward and likely misunderstood at best, we came up with the following simple protocol for installing a generic nanotechnology solution:
modifications of individual cells to express bio-molecular recorders using a lentivirus or alternative viral vector and recombinant-DNA-engineered payload,
distribution of OPID devices using the circulatory system and, in particular, the capillaries in the brain as a supply network with steps taken to circumvent the blood-brain-barrier, and
the use of isopropyl β-D-1-thiogalactopyranoside (IPTG) “subroutines” to control gene expression for molecules used in “pairing” neurons with their associated reporter OPID devices.
It is worth noting that a more natural method of “installation” might indeed involve a developmental process patterned after the the self-assembly processes that govern embryonic development, but thus far our understanding of molecular self assembly is limited to the simplest of materials such as soap films and even in these cases nature continues to surpass our best efforts.
Relating to our discussion of the steps in the above protocol, Biafra sent around the following useful links:
protecting macromolecules: PEGylation, liposomes, doxil is a drug example, etc;
conjugating macromolecules and nanoscale devices: review article, nanopores, several lab-on-a-chip type technologies (see 1, 2, 3, 4);
adapting molecular imprinting to nanoscale devices: (see this paper);
linking biological molecules: Streptavidin, Biotin, “click chemistry” (see this review);
Apropos of Wednesday’s assignment to read [22], you might want to check out this recent article [139] in Nature on Kark Deisseroth. Here are a couple of quotes from Mark Schnitzer’s papers on their fluorescence micoendoscopy technology relevant to this afternoon’s class discussion:
“The Schnitzer group has recently invented two forms of fiber optic fluorescence imaging, respectively termed one- and two-photon fluorescence microendoscopy, which enable minimally invasive in vivo imaging of cells in deep (brain) areas that have been inaccessible to conventional microscopy.” [13]“Gradient-index (GRIN) lenses [...] deliver femtosecond pulses up to nanojoule energies. Why is it that confocal endoscope designs do not extend to multiphoton imaging? Over SMF [single mode fiber] lengths as short as 1-10 cm, femtosecond pulses degrade as a result of the combined effects of group-velocity dispersion (GVD) and self-phase modulation (SPM).” [78]
On a completely unrelated note, I was impressed with Daniel Dennett’s comments about consciousness following his recent talk at Google discussing his new book entitled “Intuition Pumps and Other Tools for Thinking”:
“I do not think that [consciousness] is a fundamental category that splits the universe into two. I think the idea that it is is deeply rooted in culture and may be right, but I think that consciousness is like reproduction, metabolism, self repair, healing, blood clotting; It is an interestingly complex, deeply recursive biological phenomenon [...] I think that [human consciousness] is software, it has to be installed the implication being that [...] If a human baby could be raised completely cut off from human contact, without language, and could grow to adulthood without the benefit of all the thinking tools that would normally be installed in the course of just growing up in a social community, then I think that human being would have a mind so profoundly different from ours that we would all agree that human being wasn’t conscious [in the human sense].”
May 19, 2013
Biafra sent along these examples of computations performed in biology that hint at what might be mined from detailed studies of biological organisms [145, 80]. That each paper examines a different algorithmic approach on a different computational substrate underscores the fact that we need to be open minded when looking for computation in biology [3, 135] and creative in coming up with applications that make sense. The tools that were used in extracting these programming pearls (the title of a book and a column for the Communications of the ACM magazine written by Jon Bentley) are in stark contrast with those available to Francis Crick, Sydney Brenner, George Gamow and others trying to figure out how DNA eventually produced protein. I went back and read the relevant sections of Horace Freeland Judson’s The Eighth Day of Creation [77] to get some feeling for what it was like to unravel this puzzle using the available technology and what was known at that time.
Today we know about messenger and transfer RNA. We know that amino acids are coded as triplets consisting of three bases drawn from {A,C,G,U}, the same bases as DNA except uracil (U) replaces thymine (T). We know the code for amino acids is redundant and includes (3) stop codons, and that the remaining (61) coding codons specify twenty essential amino acids. But when Brenner, Crick and Gamow were trying to decipher the code they knew comparatively little about the molecular structure of proteins and the details of cellular biochemistry. Communication was more often than not by snail mail, made all the more painful with Brenner in South Africa, Crick in Cambridge and Gamow and Watson flitting back and forth between the UK and the East and West coast in the US. Bench scientists were inventing the reagents and protocols we take for granted today and while X-ray crystallography was relatively advanced it had limitations and you first had to know what you wanted to look at — they had to deduce the existence of ribosomes and infer the number of bases per codon.
May 15, 2013
Akram Sadek talked about his work [133] on multiplexing nanoscale biosensors using piezoelectric nanomechanical resonators. Audio for the class discussion is on the course calendar page and Akram promised to send his slides. Between the audio and slides and the three papers assigned for class I think you have plenty of background to understand his multiplexer technology in some detail. I think we were all intrigued with his description of how carbon nanotubes conjugated with DNA to render them biocompatible might be embedded in cell membranes and used as voltage sensors and to excite and inhibit individual neurons. Moreover the prospects to carry out interventions using RF rather than light as in the case optogenetics offers the potential to sense and intervene deep within tissue and without drilling holes in the skull.
Note that there is quite a bit of work on using nanotubes for drug and probe delivery. Wu and Philips [167] describe the development of single-walled nanotubes as a carrier for single-stranded DNA probe deliver. They claim their method offers superior biostability for intracellular applications including protection from enzymatic cleavage and interference from nucleic acid binding proteins. Their study “shows that a single-walled carbon nanotube-modified DNA probe, which targets a specific mRNA inside living cells, has increased self-delivery capability and intracellular biostability when compared to free DNA probes. [The] new conjugate provides significant advantages for basic genomic studies in which DNA probes are used to monitor intracellular levels of molecules.” Ko and Liu [85] report work on organic nanotubes assembled from single-strand DNA used to target cancer treatment.
I suggested the idea of inserting thin fiber-optic cables tipped with a MEMS device attached to a prism / mirror. The MEMS device could be powered by light and controlled to point the mirror in a particular direction or to scan the surrounding tissue in the vicinity of the probe’s tip. As in the case of Ed Boyden’s 3-D probes, this arrangement could be used for both transmitting and receiving information. The fiber-optic tether is crucial given the power requirements of such a device fabricated using current technology, but it would be interesting to think about the prospects for using implantable lasers and powering the MEMS device wirelessly.
Yesterday morning, I talked with George Church about his Rosetta Brain idea. The Allen Mouse Brain Atlas is an amalgam of hundreds of mouse brains. In contrast, the Rosetta Mouse Brain would combine four maps of a single mouse: connectomic, transcriptomic and proteomic information in addition to measurements of cellular activity and developmental lineage. George believes this could be accomplished in a year or so using his method of fluorescent in situ sequencing (FISSEQ) on polymerase colonies. He believes that the total funding required would be something on the order of $100K and that he could do it in his lab. George is on the west coast a couple of days each month and will be in the Bay area in June for a conference. He also mentioned that he, Ed Boyden and Konrad Kording have applied for BAM/BRAIN money to pursue their molecular ticker-tape work which he predicts will have it’s first proof-of-concept demonstration within a year.
May 13, 2013
In today’s class, we discussed class projects and, in particular, the proposals that are due next Monday at noon. Note that the due date has changed; it seems that not all of you keep late night hours and so I extended the deadline to noon. This entry will be a hash of follow-up items from our class discussion. Note that I expect your proposals to reflect the original list of top-ten promising technologies and the comments from our invited speakers and technical consultants.
First off, here are some links relevant to BAM/BRAIN that might be useful in understanding how the initiative is viewed from the perspective of two key federal agencies, NIH and NSF. And, apropos our discussion of commercial applications of BAM/BRAIN technologies, here’s an example of an interesting company in the BCI space. Ariel Garten, the CEO of InterAxon spoke in Ed Boyden’s Neurotechnology Ventures course at MIT last Fall. The title of his presentation was “Thought-controlled computing: from popular imagination to popular product.” Here’s their website splash page:
 |
Relating to Mainak and Ysis’ project focus, Justin Kinney is a good contact in Ed Boyden’s group working on strategies for neural signal acquisition and analysis. At the Salk Institute, he worked on three-dimensional reconstructions of brain tissue from serial-section electron microscopy images, and his dissertation at UCSD involved the development of software for carrying out accurate Monte Carlo simulations of neuronal circuits (PDF). This last is relevant to some of Nobie’s interests.
Also relevant to cell-body segmentation is the work of Viren Jain [74] who worked with Sebastian Seung at MIT and collaborated with Kevin Briggman and the Denk lab. Viren is now at HHMI Janelia Farm and recently gave a talk at Google that you might find useful to review along with Kevin’s presentation available from the course calendar page. You can find more recent papers at Viren’s lab at Janelia and Sebastian’s lab at MIT. The relevant algorithms involve machine learning and diverse techniques from computer-vision including superpixel agglomeration and superresolution.
Doing back-of-the-envelope calculations will play a key role in your course projects. For a classic example of a back-of-the-envelope calculation, see Ralph Alpher and George Gamow’s analysis of how the present levels of hydrogen and helium in the universe could be largely explained by reactions that occurred during the “Big Bang” (VIDEO). I mentioned Chris Uhlik’s back-of-the-envelope analysis of using RF for solving the readout problem: his analysis short, to the point, and worth your time to check out; you can find his notes here.
I had wanted to do an exercise in class in which we scale the human brain to the size of the Earth, but we ran short of time. The intent was to have you do all of the work, but I had some rough notes to help grease the wheels that I include below. I started with some comments about where you can find quantitative information. In cases where there is an established engineering discipline various companies and professional societies often provide resources, e.g., the BioSono website provides tissue-density tables, transducer simulations and a host of other useful tools and data for the engineer working on medical ultrasound imaging. In neuroscience, academic labs often compile useful lists of numbers that neuroscientists find useful to keep in mind, e.g., see Eric Chudler’s list at University of Washington.
As a concrete example, dendrites range in diameter from a few microns at the largest (e.g. the primary apical dendrite), to less than half a micron at the smallest (e.g. terminal branches). The linear distance from the basal end to the apical end of the dendritic tree usually measures in the hundreds of microns (range: 200 μm to over 1 mm). Because dendrites branch extensively, the total dendritic length of a single neuron (sum of all branch lengths) measures in centimeters.3 Now let’s see how we might use this information in our exercise to rescale the brain to be around the size of the Earth.
The Earth is about 12,742 km in diameter, which we’ll round off to 10,000 kilometers. Let’s say a human brain is 10 cm or 0.1 m on a side and so our scale factor is 100,000,000:1. The diameter of a neuron (cell body) is in the range of 4-100 μm [granule,motor] and so let’s call it 10 μm and its dendritic span could be more than a millimeter. In our Earth-size scaled brain, the diameter of a neuron would be around 1 kilometer and its axonal and dendritic processes could span ten times that or more. So now we have a neuron the size of a small town and its axonal and dendritic processes spanning a small county. What about smaller entities?
A molecule of glucose is about 0.5 nm and a typical protein around 5 nm. Synaptic vesicles can contain upwards of 5,000 molecules of small-molecule neurotransmitter like acetylcholine. Think of these relatively small molecules as being on the order of a meter in size. Bigger than a loaf of bread and smaller than a 1969 Volkswagen bug. Recall that some cellular products are manufactured in the soma and transported along microtubules to be used in the axons. What would that transport system look like?
The outer diameter of a microtubule is about 25 nm while the inner diameter is about 12 nm. These tubular polymers of tubulin can grow as long as 25 micrometres and are highly dynamic. Think of the outer diameter of the microtubule being around the width of a small-gauge railroad track. What about even smaller entities? Ions vary in size4 depending on their atomic composition and whether they have a negative or positive charge, but typical values range from 30 picometers (1012 m) (0.3 Ångström) to over 200 pm (2 Ångström). In our planet-sized brain, ions are on the order of a millimeter. Still visible but just barely.
Intracellular fluid, the amount of water that’s inside our cells, accounts for something between 2/3rds and 4/5ths of our total volume, and so it follows that the extracellular fluid, the amount of water that surrounds our cells, accounts for between 1/5th and 1/3 of our total volume. Approximately 78% of the brain consists of water. The rest is comprised of lipids, proteins, carbohydrates, and salts. The ratio of grey to white matter — which consists mostly of glial cells and myelinated axons — is about 1.1 in non-elderly adults; myelin is about 40% water; the dry mass is about 70-85% lipids and about 15-30% proteins.
The inside of cells is crowded with molecules moving around due to the forces of Brownian motion, diffusion, and the constant making and breaking of covalent bonds that serve alter molecular shapes. Some compartments and organelles are more congested than others, but these forces dominate in the propagation of information and its physical manifestation — proteins, ion concentrations — in the central nervous system. Try to imagine what this world would look like if you could walk through our Earth-size brain and watch DNA polymerase at work replicating DNA in the cell nucleus or signal conduction in a synapse.
In our planet-sized brain, we can imagine using communication satellites and cellular networks to transmit and receive information, cell phones and RFID devices using wireless, bluetooth and NFC (near-field communication) for local information processing and intra-cellular communication. It is an interesting exercise to consider Chris Uhlik’s back-of-the-envelope analysis assuming for the purpose of the exercise that your readout technologies could use the macroscale physics of a planet-sized brain while the massive brain would miraculously continue to operate using microscale physics of biology. It’s a stretch, but a good exercise.
Apart from preparing for the above thought experiment which we didn’t even end up talking about in class, I also wrestled a bit more with how to frame the reporting problem and the technologies that you’ll be investigating in your projects. In the spirit of writing sections of the paper many times in preparing for the final version, here are some of my thoughts: Without getting too abstract, energy and information are at the heart of what we’re up against; positively in terms of powering recording and reporting technologies and defining informational targets, and negatively in terms of compromising tissue health and producing noise that interferes with signal transcoding and transmission.
One way or another, we have to (a) expend energy to measure specific quantities at specific locations within the tissue, (b) expend energy to convert raw measurements into a form suitable for transmission, (c) expend energy to transmit the distilled information to locations external to the tissue, and, finally, (d) expend energy to process the transmitted information into a form that is useful for whatever purpose we have in mind, whether that be analyzing experimental results or controlling a prosthetic. Each of these four steps involves some form of transduction: the process of converting one form of energy into another.
In the case of imaging, typically all of the required energy comes from sources external to the tissue sample. Electromagnetic or ultrasonic energy is used to illuminate reporter targets and then reflected energy is collected to read out the signal. We can complicate the picture somewhat by using the energy from illumination to initiate cellular machinery to perform various information processing steps to improve the signal-to-noise ratio of the return signal. We can go a step further and use the illumination energy to power more complicated measurement, information-processing and signal-transmission steps.
Alternatively, we can figure out how to harness cell metabolism and siphon off some fraction of the cell’s energy reserves to perform these steps. In the case of external illumination, the tissues absorb energy that has to dissipated, as thermal energy. In the case of depleting existing cellular energy, the brain already uses a significant fraction of the body’s energy and operates close to its maximum capacity. In either case, if we are not careful, we can alter the normal function of the cell thus undermining the whole endeavor.
The human brain, at 2% of body mass, consumes about 20% of the whole body energy budget, even though the specific metabolic rate of the human brain is predictably low, given its large size. Source: Herculano-Houzel, S. Scaling of Brain Metabolism with a Fixed Energy Budget per Neuron: Implications for Neuronal Activity, Plasticity and Evolution. PLoS ONE (2011). The brain’s use of glucose varies at different times of day, from 11 percent of the body’s glucose in the morning to almost 20 percent in the evening. In addition, different parts of the brain use different amounts of glucose.
Several factors make it difficult to identify specific metabolic requirements. First, we know the brain is constantly active, even at rest, but we don’t have a good estimate of how much energy it uses for this baseline activity. Second, the metabolic and blood flow changes associated with functional activation are fairly small-local changes in blood flow during cognitive tasks, for example, are less than 5 percent. And finally, the variation in glucose use in different regions of the brain accounts for only a small fraction of the total observed variation. Source: Raichle, M.E., Gusnard, D.A. Appraising the brain’s energy budget. Proceedings of the National Academy of Science (2002). (PDF)
May 11, 2013
There are eight students taking the course for credit. I took the initial list of proposed top-ten promising technologies and tried to group them in related pairs. We’ll revisit this grouping during class discussion on Monday, in preparation for which you should also review the comments from our invited speakers and technical consultants. I ended up with four pairs and two singletons and then I made a first pass to assign each of you to one of these six categories.
All of this preliminary organization is up for discussion and indeed whatever technologies you end up taking responsibility for in your final project and the proposed technical report, I expect that you will consult with each other on other technologies depending on your particular expertise. Here are my initial assignments: (#1 and #5) piggybacking on medical imaging — Anjali, (#2 and #6) finessing imaging challenges — Nobie, (#4) implantable and steerable arrays — Daniel, (#3 and #8) robotics and computer science — Mainak and Ysis, (#7) leveraging genome sequencing — Biafra, and (#9 and #10) advances in nanotechnology — Oleg and Andre. And here is a simplified version of the previously circulated list aligned with the above-mentioned indices for easy reference:
near-term advances in imaging for human studies — MRI, MEG, FUS, PAI, PET, SEM, etc. — (1-2 years)
development of new animal models — transgenic fly, mice, zebrafish, etc. – (2-3 years)
automation using machine learning and robotics — electron microscopy, animal experiments, etc. — (1-2 years)
advanced fiber-optically coupled multi-cell recording — coupled 3-D arrays, microendoscopy — (2-3 years)
intermediate-term advances in imaging broadly construed — MRI, FUS, SEM, etc — (2-3 years)
new contrast agents and staining preparations — MRI, CLARITY, ultrasound — (2-3 years)
leveraging advances in genome-sequencing — DNA barcoding, molecular ticker-tapes — (3-5 years)
computation-intensive analytics — cell-body segmentation, spike-coding, simulations — (1-2 years)
semiconductor-based recording with optical readout — quantum dots, FRET technology, etc — (3-5 years)
implantable bio-silico-hybrids, wireless or minimally-invasive fiber-optic readout — (5-10 years)
Miscellaneous topics that might be worth addressing in the technical report:
Translational medicine: mouse and human studies and investment in the technologies that support them;
Evolution of mature technologies and how to accelerate development of promising nascent technologies;
Technological opportunities and predicted time-lines for the longer-term impact of nanotechnology;
Potential for significant infusion of investment capital to spur innovation akin to genome sequencing;
I’m often chided for having a “reductionist” philosophy in terms of my scientific and technological outlook, and so I enjoyed the following excerpt from The Rapture of the Nerds: A tale of the singularity, posthumanity, and awkward social situations by Cory Doctorow and Charles Stross. Huy, the book’s protagonist, is an uploaded human in a post-singularity era in which most of the humans and all of the “transcended” humans exist in simulated worlds that are run on a computational substrate constructed from extensive mining of the solar system.
Just prior to the exchange described in the excerpt, she — Huy started the book as a flesh-and-blood human male but was slotted into a female simulacrum in the upload process — asks her virtual assistant referred to here as the “Djinni” how to adjust her emotional state and is presented with an interface replete with cascading selection menus and analog sliders of the sort you might find in a studio recording mixer.
The book is full of obscure science fiction references, and the following excerpt refers to a fictional device called a “tasp” that induces a current in the pleasure center of the brain of a human at some distance from the wielder of the tasp. In Larry Niven’s novel Ringworld, these devices are implanted in the bodies of certain members of an alien race known as Puppeteers for the purpose of conditioning the humans they have to deal with in order to render them more compliant. Here’s the excerpt:
“I hate this,” she says. “Everything it means to be human, reduced to a slider. All the solar system given over to computation and they come up with the tasp. Artificial emotion to replace the genuine article.”The Djinni shakes his bull-like head. “You are the reductionist in this particular moment, I’m afraid. You wanted to feel happy, so you took steps that you correctly predicted would change your mental state to approach this feeling. How is this different from wanting to be happy and eating a pint of ice cream to attain it? Apart from the calories and the reliability, that is. If you had practiced meditation for decades, you would have acquired the same capacity, only you would have smugly congratulated yourself for achieving emotional mastery. Ascribing virtue to doing things the hard, unsystematic way is self-rationalizing bullshit that lets stupid people feel superior to the rest of the world. Trust me, I’m a Djinni: There’s no shame in taking a shortcut or two in life.”
May 9, 2013
Yesterday’s class discussion with Brian Wandell was timely. Brian started by making a case for more emphasis on human studies. First, he claimed that only 1/3 of 1% of mouse studies result in human drug or clinical-treatment trials. Without data on how many human studies result in such trials, this statement is a bit hard to put in perspective, but it is sobering nonetheless. It would also be useful to have statistics on how often a promising molecule or intervention found in a study involving a mouse model translates to a corresponding result in humans. I asked Brian for relevant papers and he suggested two [62, 153] that look promising.
To underscore the differences between human and mouse models, Brian mentioned that myelinated axon tracts in the mouse brain are far less common than in human brains — or, more precisely, the fraction of total brain volume occupied by white matter is significantly less in mice — in part because the distances involved in the mouse are considerably shorter and hence faster signal conduction is less advantageous. He also mentioned differences in the visual systems and retina; human eyes converge to focus and have high acuity in the foveal region while mice tend to diverge to gain greater field of view and their retinal cells exhibit less diversity tending to behave for the most part like the human retinal cells responsible for our peripheral vision. This reminded me of Tony Zador’s comment that humans and more highly-evolved organisms tend toward more specialized functions facilitated by complicated genomic pathways involving more promoters and switches than the corresponding protein-expression pathways in simpler organisms.
Having motivated the study of human brains, he turned his attention to one of most powerful tools for studying human cognition. Brian summarized how fMRI is employed in studying cognitive function in humans, including normal and pathological behaviors involving perception, language, social interaction, executive function, affective disorders including anxiety and depression, and developmental aspects of the aforementioned. I promised to include a pointer to the work of Stanislas Dehaene and so here are some of Dehaene’s technical papers and books [47, 46, 44, 45], the last of which Reading in the Brain: The Science and Evolution of a Human Invention is an excellent introduction to the research on reading that Brian talked about in his presentation.
Brian also provided background on the basic physics of NMR, how it’s applied in the case of diffusion tensor imaging (DTI), and how his lab is attempting to improve resolution and extend the class of neural signals it is possible sample with this technology. The multi-plane parallel-imaging technology5 soon to be available will accelerate imaging four-fold, and perhaps there are even greater gains to be had in the future. Speed definitely matters in both clinical and scientific studies. A friend of mine just recently had to endure an MRI scan that required him to remain still for 45 minutes and would have been very grateful had Stanford employed the new technology Brian told us about. Biafra Ahanonu recommended this website maintained by Joseph Hornak at the University of Rochester as a convenient resource and introduction to MRI.
At the workshop sponsored by The National Science Foundation & The Kavli Foundation entitled “Physical and Mathematical Principles of Brain Structure and Function”, Huda Zoghbi spoke (PDF) eloquently about the need to “know all of the components of the brain, not just neurons” and that we need know how these components connect and communicate and most important “how brain activity varies with experiences and external stimuli.” She points to the Allen Institute and the foundation it has established in developing the paradigms and open-source resources for the Mouse Brain Atlas. I found her discussion of the MeCP2 gene instructive in light of Brian’s comments about mouse studies. MeCP2 causes Rett syndrome whose symptoms range from loss of language and social skills around two years of age to inability to control movement. I think one could argue that preliminary effort expended on MeCP2 in mouse models was well spent, if it ultimately assists in human studies.
She opines that “biology, genome projects, and neuroscience have taught us the value of the model organism” and that “we need to continue to capitalize on this.” And then a little later, citing the work of Helen Mayberg who used MRI analysis of white matter to study the difference between patients suffering from depression who respond to deep brain stimulation and those who don’t, she suggests that “somehow we need to make the research more iterative between humans and the lab.” Elsewhere Zoghbi posits that it is hard to argue that “technology development wasn’t a major part of the genome project” citing Ventner’s use of genome-wide shotgun assembly which “came mostly from improvements in algorithms, longer read lengths and computational power” and observes that the “benefits of the influx of bioinformatics due to the genome project have spread well beyond genome science. She suggests to the participants that we need to “think about what technology needs to be developed [for BAM/BRAIN], and what portion of [NSF] funding needs to be set aside for technology development.”
In this interview with Sydney Brenner — see Part 1 and Part 2, Brenner called Craig Ventner’s synthetic organisms “forgeries” likening them to the Vermeer forgeries by Han van Meegeren, and expressed his belief that we’re drowning in data and starving for knowledge — paraphrasing John Naisbitt in his 1982 book Megatrends, and that all this Bayesian analysis and cataloguing was so much bean counting and stamp collecting without meaningful theories, by which I think he meant something more akin to the Bohr or Rutherford models of the atom than to quantum electrodynamics with its reliance on probabilistic wave functions.
Similar sentiments were expressed by the participants of the workshop mentioned earlier. On the first day of the workshop all of the participants were given the opportunity to express their interests relating to BAM/BRAIN in one minute or less. The audio for those sessions are available here and here if you’re interested. No neuroscientist in this day and age is going to say publicly that theory does not deserve a central role in the brain sciences — the words “hypothesis” and “theory” often used interchangeably are inextricably linked to the scientific method. That said I think a case can be made that what constitutes a meaningful theory may have to be expanded to include new ways of thinking about the sort of complex emergent phenomena that characterize many biological and social processes.
Perhaps our craving for elegant, easily communicated theories/stories speaks to inherent limitations in our cognitive capacity. The search for general principles — the holy grail of modern science — sounds so reasonable until you ask scientists what would qualify for such a principle, and here I suspect you’d get very different answers depending on whom you ask. For example, would Donald Hebb’s proposed mechanism for synaptic plasticity or Francis Crick’s central dogma of molecular biology pass muster with most neuroscientists if they were proposed today? We would like the world to be simple enough accommodate such theories, but there is no reason to expect that nature or the cosmos will cooperate. Perhaps in studying complex systems like the brain, ecosystems or weather, we’ll have to settle for another sort of comprehension that speaks more to the probabilistic interactions among simpler components and characterizations of their equilibrium states without a satisfying account of the intermediate and chaotic annealing processes that seek out these equilibria.
Moreover, complex emergent phenomena like those produced by living organisms can only fully be explained in the context of the larger environment in which the relevant systems evolved, e.g., adaptations to a particular constellation of selection pressures. In some circles, the term “emergent” is often use derisively to imply inexplicable, opaque or even mystical. As for “stamp collectors”, the modern day scientific equivalent of the naturalists who collected species of butterflies or ants in the Victorian age, consider Craig Ventner’s sailing the world’s oceans in search of new organisms, molecular biologists looking for useful biomolecules, e.g., channelrhodopsin, to add to their toolkit for manipulating life at the nanoscale, and virologists on the quest to discover the next interesting microbe. And I would add another potentially fruitful avenue for future exploration. Why not search for useful computational machinery as revealed in cellular circuits (not just neurons) to solve algorithmic problems on computational substrates other than biological ones?
May 7, 2013
In motivating faster, less expensive methods for extracting the connectome, Tony Zador offered several scientific assays that connectomes would make possible. For example, he described work by Song et al [140] from Chklovskii’s lab analyzing how local cortical circuitry differs from a random network. They discovered that (a) bidirectional connections are more common than expected in a random network and (b) that certain patterns of connectivity among neuron triples were over represented. These experiments were extremely time-consuming and Tony pointed out that considering patterns involving more than three neurons would be prohibitively time consuming. He also noted that large-scale spectral graph analysis would be feasible with a complete connectome and that finding patterns such as those revealed in the Song et al work could be carried out easily with the connectome adjacency matrix.
Tony then described the core idea — recasting connectomics as a problem of high-throughput DNA sequencing, his inspiration — Cold Spring Harbor is a world-renowned center for the study of genomics and he had sat through many talks by visitors discussing sequencing and analyzing the genomes of various organisms, and, finally, his education in molecular biology and its acceleration during conversations with his running partner Josh Dubnau, a molecular biologist and colleague at Cold Spring Harbor lab. We had already covered the basics — a multi-step process consisting of (a) barcoding individual neurons, (b) propagating barcodes between synaptically-coupled neurons, (c) joining host and invader barcodes and (d) sequencing the tissue to read off the connectome — from reading his earlier paper [172], and were primarily interested in progress on those problems that were not adequately addressed in that earlier paper.
He began with a review of the earlier work and outlined his strategy for validating his method. His ultimate goal is the mouse connectome — ~105 neurons, not all stereotyped and roughly as complex as a cortical column, but his interim goal is to sequence the connectome of C. Elegans — 302 neurons and ~7,000 stereotyped connections — for which we already have ground truth thanks to the heroic work of Sydney Brenner6 and his colleagues [165]. This two-organism strategy introduces formidable challenges as his technique must apply to both a mammal and a nematode with different neural characteristics and, in particular, different susceptibility to viruses of the sort Zador proposes to use to propagate information between neurons.
He talked briefly about adding locality and topographic annotations to the connectome. Some locality will fall out of slicing and dicing the tissue into 3-D volumes a few microns on a side to be sequences individually thereby associating barcoded neurons with locations in 3-D space, but he also mentioned the idea of barcoding neuron-type information thereby labeling individual neurons and sequencing this along with the barcoded connectomic information. In response to a related comment by Adam Marblestone in an earlier post, I sketched Tony’s proposed method and initial proof-of-concept experiments — see here.
Tony then launched into a discussion of barcoding beginning with the basics of using restriction enzymes to cut a plasmid vector at or near specific recognition nucleotide sequences known as restriction sites in preparation for inserting a gene or, in our case, a barcode. He first described the relatively easy case of in vitro cell cultures and then took on the challenge of in vivo barcoding which is complicated by the requirement that the random barcodes must be generated in the cells. Given a standard template corresponding to a loxP cassette, the goal is to randomize the template nucleotide sequence by applying one or more operators that modify the sequence by some combination of insertion, deletion, substitution and complementation operations.
Cre recombinase, a recombinase enzyme derived from a bacteriophage, is the tool of choice for related applications. Unfortunately Cre performs both inversion — useful in randomizing a sequence — and excision — not so useful as it shortens the sequence yielding an unacceptable O(n) level of diversity : “DNA found between two loxP sites oriented in the same direction will be excised as a circular loop of DNA whilst intervening DNA between two loxP sites that are oppositely orientated will be inverted.” The solution was to find a recombinase that performs inversion but not excision.
Rci recombinase, a component in the aptly named shufflon inversion system, was found to fill the bill yielding a theoretical diversity of O(2n n!) and an in vivo experimental validation that more than satisfied their requirements. Employing a model in which a lac promoter was used turn Rci expression on or off, they ran a control with Rci off resulting in no diversity and then a trial with Rci turned on that produced the desired — exponential-in-the-length-of-the-template — level of diversity. The initial experiments were done in bacteria. Rci also works in mammals and subsequent experiments are aimed at replicating the results in a mouse model.
Next Zador provided some detail on how they might propagate barcodes transynaptically using a pseudorabies virus (PRV). PRV spreads from neuron to neuron through the synaptic cleft. The PRV amplicons are plasmids that include a PRV “ORI” sequence that signals the origin of replication and a PRV “PAC” sequence that signals what to package. They can be engineered so that they don’t express any viral proteins thus rendering them inert at least in isolation. A helper virus is then used to initiate synaptic spread in a target set of neurons.
The in vivo experiments were reminiscent of the circuit-tracing papers out of Ed Callaway’s lab, dealing, for example, with the problem of limiting spread to monosynaptic partners — see for example [30, 105]. Researchers in Tony’s lab have replicated Callaway’s rabies-virus experiments using their pseudorabies virus, and so there is ample precedent for the team succeeding with this step in the overall process. The final step of joining host and invader barcodes into a single sequence was demonstrated in a very simple experiment involving a culture of non-neuronal cells and then in another in vitro somewhat more complicated proof-of-concept experiment.
In this second experiment, two sets of embryonic neurons were cultured separately, one with the PRV amplicon alone and a second with PRV amplicon plus the φ31 integrase enzyme which was introduced to mediate the joining of host and invader barcodes. Then the two cultures were co-plated and allowed to grow so as to encourage synaptic connections between neurons, the cells are infected with PRV thus initiating transynaptic transfer of barcodes between connected neurons, and, finally, after a sufficient interval of time to allow the connections to form, the sample is prepared, the DNA amplified by PCR and the result sequenced. Without accurate ground-truth, it is impossible to evaluate the experimental quantitatively, but qualitatively the resulting connectome looked reasonable.
There is still some way to go before the Zador lab is ready to generate a connectome for C. Elegans, but they have made considerable progress since the 2012 paper in PLoS in which they proposed the basic idea of sequencing the connectome. Tony also talked about alternative approaches, and we discussed how, as in the case of optogenetics, a single-molecule solution might surface that considerably simplifies the rather complicated process described above. The prospect for serendipitously stumbling on the perfect organism that has evolved an elegant solution to its survival that also solves a vexing engineering problem is what keeps molecular biologists avidly reading the journals of bacteriologists, virologists and those who study algae and other relatively simple organisms that exhibit extraordinarily well adapted survival mechanisms.
May 5, 2013
In response to my request for feedback regarding our initial list of promising technologies to focus on for the remainder of the course, I received a number of very helpful replies, several of which are highlighted here:
Bruno Olshausen mentioned that traditional methods for recording electrical activity — which in his view is still the gold standard — including patch-clamping and 2-D and 3-D arrays, are improving all the time. He mentioned two technologies we might look at more carefully: The first involves the use of polytrodes that Ed Boyden mentioned in his survey of microscale recording technologies. Polytrodes provide a quality of single-unit isolation that surpasses that attainable with tetrodes. Bruno suggested we look at the work of Nick Swindale and Tim Blanche [19]. He also mentioned that “Charlie Gray has been using this methodology to record activity across all layers of cortex simultaneously in response to dynamic natural scenes, the data you get from this are stunning in terms of providing new insights about neural dynamics.” The second technology Bruno mentioned involves electrocorticogram (ECoG) arrays that provide local field potentials (LFPs) at high density across an area of cortex. Bruno writes that:
Bob Knight and Eddie Chang’s lab in particular have been making some amazing findings with these arrays, showing you can decode speech from patterns of activity over auditory cortex. I think it speaks to the fact that many of the response properties we see in individual neurons actually reflect more macroscopic variables carried by the entire population. I believe the finer grained aspects of activity at the individual neuron level are probably carrying much more advanced and complex forms of information that we as yet just don’t have the intellectual maturity (or perhaps capacity) to appreciate. That’s not to say we shouldn’t record at that level, but we should at least be aware that the stories we make about tuning properties and so forth of individual neurons is just nonsense.ECoG arrays are an invasive technology but one that appears to be relatively well tolerated by the implanted animals. Rubehn et al [131] describe a 252-channel ECoG-electrode array made of a thin polyimide foil substrate enclosing sputtered platinum electrode sites and conductor paths that subtends an area of approximately 35mm by 60mm. In studies involving implantation in Macaque monkeys, all electrodes were still working after 4.5 months with no discernible decline in signal quality.
Mike Hawrylycz mentioned that single-cell gene-expression profiling techniques [96] are becoming very sophisticated and offer interesting insights into the dynamics of individual neurons:
A key goal of biology is to relate the expression of specific genes to a particular cellular phenotype. However, current assays for gene expression destroy the structural context. By combining advances in computational fluorescence microscopy with multiplex probe design, we devised technology in which the expression of many genes can be visualized simultaneously inside single cells with high spatial and temporal resolution. Analysis of 11 genes in serum-stimulated cultured cells revealed unique patterns of gene expression within individual cells. Using the nucleus as the substrate for parallel gene analysis, we provide a platform for the fusion of genomics and cell biology: “cellular genomics.” Source: Levsky, J.M. and Shenoy, S.M. and Pezo, R.C. and Singer, R.H. Single-cell gene expression profiling. Science. 2002 Aug 2;297(5582):836-40.
George Church mentioned his project to use fluorescent in situ sequencing (FISSEQ) on polymerase colonies [111] (polonies) in which they expect to be able to integrate in a single brain five key measurements at cell/subcellular resolution, including connectomic, transcriptomic7 and proteomic information in addition to measurements of cellular activity and developmental lineage. He claims that FISSEQ offers a nearly infinite number of colors (460) and hence should enable highly accurate circuit tracing with 300nm imaging rather than 3nm resolution, resulting in a million times less raw data to deal with.
TLD: The word “polony” is a contraction of “polymerase colony,” and corresponds to a small colony of DNA. Polonies are discrete clonal amplifications — typically obtained using some variant of the polymerase chain reaction (PCR) method — of a single DNA molecule, grown in a gel matrix. Polony sequencing refers to a method of that uses multiplex assays to achieve high accuracy with multiple reads run in parallel. The use of polonies in gene sequencing significantly improves the signal-to-noise ratio. (source)
Here is some feedback from Adam Marblestone:
I like the focus on “scalable neuroscience” which is a fairly broad concept.
I think you should distinguish more clearly between structural / anatomical mapping and dynamic activity mapping. These support each other, and there are potential hybrids — like Zador-barcoded tickertapes — but they are quite distinct goals, and some prominent scientists believe strongly in the desirability of one goal but not the other.
Regarding footnote #3: Didn’t Viren presentation mention that their focused ion beam milling approach at Janelia can allow z slices of something like 5 or 10nm at this point? That would seem to totally solve this problem.
Regarding George’s idea of using in-situ sequencing to get a near-infinite number of digitally-distinguishable cell “colors” (barcodes) for structural connectomics, I wonder if this really solves the problem: can you detect synapses when operating at the 200-300nm resolution limit of diffraction-limited optics? The problem is to distinguish synapses from close appositions of cell membranes that are not actual synapses. One way around this is to directly label synapses using GRASP, or various antibody labels. These could be adapted to super-resolution optical microscopy labels (which photo-switch or blink) so as to see almost every synapse and to localize the synaptic contacts to maybe 20nm precision or so, although at a big cost to the imaging speed. But you also have to say which sequence-barcoded cells are synapsing at any given synapse: even with in-situ sequencing and a separate synapse-specific label, that could be challenging — the cells are so densely packed and the processes so thin and tangled, which is why the ~1nm resolution of EM has been considered to be crucial.
Also, how this works depends on the density of the polonies scattered throughout the cell — you could potentially miss fine dendritic or axonal processes just because there is no polony that grew there.
It might be interesting to think about combining this with in-situ readout of Zador barcodes, which would more directly give you information about DNA exchange across synapses. Regarding viral barcodes, though, one issue is that the trans-synaptic transfer is not 100% efficient, and it is also not yet certain that the viruses only cross at synapses, not at close membrane-membrane appositions which are not synapses.
These DNA + optical readout technologies for anatomical / molecular connectomics can be comparatively easily combined, in principle. Whereas EM will see every synapse and fully trace the membranes, but is harder to combine with other techniques.
TLD: Tony Zador described a clever way to hijack RNA splicing in order to tag mRNAs with the host barcodes which are subsequently sequenced to yield cell-type signatures. He told us about a proof-of-concept demonstration in which they succeeded in an in vitro experiment using human embryonic kidney (HEK) cells. They observed high rates of premature cell death in the HEK cultures, but Tony believes this was because the embryonic cells were overly aggressive initiating apoptosis at the least sign of replication errors. Adult neurons seldom replicate and hence they can afford to be less fussy with respect to replication errors.
“Here are possibilities that might serve as a start: testing theories relating to the function of retinal and cortical circuits, e.g., Marcus Meister’s work on direction selectivity in the retina [17] and Clay Reid’s work on preferred stimulus orientation in mouse visual cortex [4] (1-2 years), using direct optogenetic control and feedback to implement interventions in cortical circuits for clinical purposes [26, 29, 24, 8] (2-3 years), non-invasive, portable, affordable BCI technologies for consumers and prosthetics for amputees and patients with neuro-degenerative diseases [2] (3-5 years), technology for mining neural circuits for robust machine learning algorithms (1-2 years).”
Framing these diversely-risked applications is really important. Stimulation and real-time bi-directional neural interfaces are likely to have comparatively near-term clinical and commercial applications.
May 3, 2013
The following is a note sent to the technical experts we recruited to help out with CS379C this year by advising students and providing technical consulting:
We’re at the point in the quarter when students in CS379C submit formal proposals for their class projects. This year we all agreed to work toward a collective product corresponding to a technical report surveying current and proposed technologies for addressing the readout problem. Toward that end I wrote up a short “state of our collective knowledge” document that identifies what we believe to be the most promising technologies along with a rough ordering in terms of their expected value to BAM/BRAIN, taking development time into account. The next step is to vet this list with those of you who have agreed to serve as informal advisers on the course, and then farm out the technologies — or technology areas — to the students for them to focus on for their class projects. I’d appreciate your taking a quick look at the list below and telling me if I’ve missed anything crucial or emphasized anything inappropriate. The references and footnotes are long and can be ignored, but the proposed list of candidate promising technologies is short. Thanks in advance for your input:
Objective: Assess promising technologies for scalable neuroscience with a focus on solving the readout (reporting) problem.
Here is my list of most the promising technologies sorted by return on investment focusing on near- to medium-term delivery:
advances in imaging broadly construed — largely finesse the problems relating to powering reporting devices and performing computations related to signal processing, compression, and transmission by performing all these functions external to the brain; example technologies include — MRI, FUS (focused ultrasound), PAI (photoacoustic imaging), two-photon calcium imaging, array tomography for proteomics, imunoflorescence for genomics and light-sheet fluorescence microscopy — (1-2 years)
development of new animal models exploiting characteristics of novel organisms including naturally transparent embryonic zebrafish [4], and transgenetic mice [65] and fly models enabling highly-parallel two-photon fluorescence microscopy8 — (2-3 years)
applications of machine learning and robotics to automate tasks previously carried out by scientists and volunteers, including patch-clamp electrophysiology and probe insertion [87, 38], serial block-face scanning electron microscopy, cell-body segmentation and axon tracing [24, 74], and high-throughput animal-behavior experiments [175] — (1-2 years)
advances in fiber-optically-tethered, densely-packed 3-D arrays [176, 133], and confocal, single- and multi-photon fluorescence microendoscopy [13, 84]9 — (2-3 years)
while dramatic improvements in the spatial and temporal resolution for MRI, MEG, SEM10 and diagnostic ultrasound are unlikely in the short term, incremental progress seems assured11 and substantial improvements in throughput achieved by parallel scanning techniques already show promise for MRI [56], SEM12, and ultrasound [100] — (2-3 years)
new contrast agents such as those being explored for MRI by Hsieh and Jasanoff [69], confocal microscopy by Chung et al [35], and ultrasound by Wang et al [163] — also see an earlier survey paper from Dijkmans et al [49] are likely enhance the capability of these technologies considerably — (2-3 years)
new approaches for leveraging genome-sequencing and retroviral-circuit-tracing technologies, including Zamft et al [173] for using DNA polymerase to record membrane potentials and the Zador et al [172] DNA barcoding approach for reading off connectomic information — (3-5 years)
computation-intensive analytics, e.g., sophisticated spike-sorting algorithms, and high-fidelity simulations for in silico experiments for vetting BAM/BRAIN technologies and evaluating competing hypotheses, e.g., Lucic and Baumeister [98], spike inference from calcium imaging, e.g., Vogelstein et al [160], and large-scale neural-circuit simulations13 — (1-2 years)
quantum dots, inorganic dyes and FRET technologies are promising but face toxicity challenges [29, 174, 103] — (3-5 years)
implantable very-low-power, bio-silico-hybrid chips with some form of wireless or minimally-invasive fiber-optic network for readout [52, 27, 95] — (5-10 years)
Our goal here is not to pick winners and losers but rather to identify opportunities and assess the availability of these enabling technologies over the span of the next ten years. Given that we believe these new technologies will accelerate research and enable new science, it is important to convince experimentalists of the advantages of partnering with technologists and engineers to influence the design of new scientific instruments and test out the prototypes.
Incumbents and entrenched interests always have an advantage, since a mature technology can incrementally elevate the bar long enough that nascent technologies are discouraged, finding it difficult to attract advocates and funding. By making sure that stake holders are aware of opportunities afforded by the new technologies and have realistic estimates of their delivery, we hope to engage experimentalists to help define and test new technologies, and venture capitalists to invest in them encouraged by our collective exploration of the potential both for scientific progress and timely monetization.
If successful, the report will be read by a wide variety of people and so we should give some attention to outcomes and their probable time lines to motivate investors, program managers and the general public: Here are possibilities that might serve as a start: testing theories relating to the function of retinal and cortical circuits, e.g., Marcus Meister’s work on direction selectivity in the retina [82] and Clay Reid’s work on preferred stimulus orientation in mouse visual cortex [20] (1-2 years), using direct optogenetic control and feedback to implement interventions in cortical circuits for clinical purposes [150, 164, 123, 34] (2-3 years), non-invasive, portable, affordable BCI technologies for consumers and prosthetics for amputees and patients with neuro-degenerative diseases [11] (3-5 years), technology for mining neural circuits for robust machine learning algorithms (1-2 years).
May 1, 2013
Yael Maquire introduced a number of interesting ideas in today’s class discussion. Citing the work of Richard Berkovits [15] on the sensitivity of multiple-scattering speckle patterns to the motion of a single scatterer, Yael posited the interest hypothesis that “mesoscopic scattering in tissue is an optical amplifier above body noise of 100kHz.” While considerable work would likely have to be done to figure out how to practically take advantage of Berkovits’ analysis in decoding backscatter patterns in the presence of ongoing changes in the target tissue, it is an intriguing proposal.
Yael also mentioned the efficiency of liquid crystals (greater than 95% efficient) and their potential value for neural recording devices if their resolution density could be improved, noting that consumer electronics, which has served as the main driver for LCD technology, has had no reason as yet to focus on greater resolution density.
As for in situ nanoscale devices and how they might be powered and read out, Yael provided a highly schematic design for a device consisting of a light-sensitive unit constructed from a photovoltaic cell plus an LCD reflector, a protein controller, and a CMOS logic unit consisting of ~5000-10000 transistors, with an axon-specific antibody coating. He then proceeded to describe a preliminary analysis he conducted in collaboration with researchers at the Martinos Lab using Monte Carlo simulation of light scattering and absorption in neural tissue to determine the extent of tissue in which such nanoscale devices could be powered up assuming different energy requirements.
Yael mentioned the optical window in tissue which defines the range of wavelengths where light has its maximum depth of penetration. Absorption and scattering are the primary factors limiting penetration depth in tissue. Rayleigh scattering is the elastic scattering of light by molecules and particulate matter much smaller than the wavelength of the incident light — note that visible light has a wavelength 390 to 700nm and cellular structures are composed of molecules only a few Ångströms in diameter (one Ångström equals 0.1nm).
In general, light propagating through matter can be attenuated by absorption, reflection and scattering. In the case of absorption, energy in photons is taken up by matter, converting the electromagnetic energy into internal energy of the absorber, say, in the form of thermal energy.
Reflection is the change in direction of a wavefront at an interface between two different media, which presumably applies to cell membranes, capillaries, blood plasma and intra- and extra-cellular fluids with varying concentrations of ions, nutrients and signalling proteins. In diffuse reflection, photons are absorbed and then subsequently emitted. In specular reflection, photons effectively bounce off the surface and are not absorbed and then re-radiated.
April 29, 2013
Adam Marblestone’s molecular ticker-tape slides and the audio from his discussion in class are linked to the calendar page. In addition to this line of research, Adam has also done interesting work on rapid prototyping of three-dimensional DNA origami — see this video for an introduction. Here are some of the questions that Adam addressed in class that are relevant to predicting how the ticker-tape technology might evolve and when a reasonably mature might conceivably become available to researchers:
We agreed on an initial milestone demonstrating the promise of such technology. Prepare the target organism using some combination of recombinant DNA, virus-mediated transduction, etc. Use optogenetic signalling or other suitable technology to initiate recording or, better yet, record continuously on a closed-loop template and use appropriate signalling to halt recording and mark the location on the ticker-tape. Present the organism with a stimulus — we talked about a variant of Hubel and Wiesel’s orientation-selectivity experiments [71], terminate the trial — even recording for a couple of seconds would provide a compelling demonstration, sacrifice the organism, and slice, dice, sort and sequence the ticker tapes.
We discussed properties of DNAP that would make an optimal foundation for recording, and considered how each of Taq POL (T. aquaticus), Dpo4 (S. Solfataricus) and Klenow (E. Coli) fall short, e.g., Dpo4 is slow at about 1 nucleotide per second and its misincorporation rate is sensitive to Manganese and pH, but not Calcium. Specifically, we considered the number of base pairs a DNAP will process before falling off the strand, variability in the rate of replication, the misincorporation rate and related cation dependence — Calcium is probably the best target but currently our best options involve Manganese which is not directly correlates with the signal we want to record.
Other challenges that need addressing to achieve a convincing milestone demonstration include how to start and stop the ticker-tape running so that recordings from different locations — we might also want to record from multiple sites within a single cell — are synchronized, how to attach additional barcoding so we can associate recordings with specific neurons, as well as provide connectomic and cell-type information, and how to infer cation concentrations — and the action potentials they serve as proxies for — from misincorporation rates extracted from a given ticker tape. Take a look at Adam’s ions-to-errors-baby-step slides for more detail.
There are a lot of moving parts in the ticker-tape technology. We haven’t even begun to address the questions mentioned in my posting prior to class concerning how we might package all the necessary enzymes, nucleotide recycling, and metabolic machinery necessary to initiate and sustain DNAP reactions outside of the nucleus, nor have we given more than a brief nod to the energy budget required to support recording over any but the shortest of durations. Regarding my earlier question about packaging such machinery and possibly manufacturing artificial organelles, not surprisingly there have been some initial steps in this direction worth mentioning.
I found several papers on applying what we know about organelles found in nature to build artificial organelles that could, for example, perform functions similar those carried out in the Golgi apparatus and endoplastic reticulum [106]. More interesting for our purposes, however, is recent work on compartmentalization of function involving the design of synthetic cells and organelles, e.g., see [41, 128, 53]. The following abstract from Kim and Tullman-Ercek [53] provides a nice summary of how bioengineers view the potential of developing such technologies:
Advances in metabolic engineering have given rise to the biological production of novel fuels and chemicals, but yields are often low without significant optimization. One generalizable solution is to create a specialized organelle for the sequestration of engineered metabolic pathways. Bacterial micro compartments are an excellent scaffold for such an organelle. These compartments consist of a porous protein shell that encapsulates enzymes. To re-purpose these structures, researchers have begun to determine how the protein shell is assembled, how pores may be used to control small molecule transport across the protein shell, and how to target heterologous enzymes to the compartment interior. With these advances, it will soon be possible to use engineered forms of these protein shells to create designer organelles.
April 27, 2013
As mentioned in an earlier discussion, caging is an alternative to the use of optogenetic techniques for photostimulation. Caging involves a pair of molecules (groups) consisting of a signaling molecule and a light-activated molecule. These two molecules are joined by a covalent bond that serves to suppress the activity of the signaling molecule [2]. The resulting compound is introduced into a cell and the signaling molecule activated using one- or two-photon imaging. When these compounds absorb photon(s), the caged group is cleaved and the active biological signaling molecule is released.
Calcium ions (Ca+2) and glutamates (carboxylate anions and salts of glutamic acid) are the most common signaling molecules used for photostimulation. Calcium ions are a common signaling pathway in synaptic transduction in which voltage-gated ion channels serve to release vesicles containing neurotransmitters such as acetylcholine into the synaptic cleft. Glutamate is the most abundant excitatory neurotransmitter in the vertebrate nervous system, and recall from our discussion with Stephen Smith that NMDAR — a glutamate receptor — is implicated in the control of synaptic plasticity, learning and memory formation. In vitro experiments using a combination of caged glutamate to stimulate and calcium imaging to report have proved useful in understanding synaptic transmission and circuit activity [118].
In re-reading the Zamft et al [173] in preparation for Adam Marblestone’s visit on Monday, I came up with a number of questions we might want to think about in attempting to predict of how many years of development will be necessary before a reasonably-mature ticker-tape technology is ready for prime time — think about optogenetics, CLARITY, and rabies-virus-enabled circuit tracing. I suggest we devote the latter part of our discussions with Adam and Yael to the exercise of making such predictions for the technologies they’re working on. Here are a few of the questions I came up with for Adam:
new developments concerning the choice of DNAP: Dpo4 (S. Solfataricus) and Klenow (E. Coli) — particularly the former with its acid- and temperature-tolerant origin, its relationship to Taq polymerase of PCR fame, and its odd collection of features [152];
what about thwarting the existing cellular error-correcting processes responsible for correcting polymerase replication errors;
how do we package the DNAP, the DNA “blank-tape” template and any additional machinery required to handle the simplest variant of direct recording on the DNA template;
how do we produce the necessary energy and raw materials and what is the metabolic cost of synthesizing a supply of nucleotides for polymerase-enabled recording;
perhaps we could package the high-misincorporation-rate DNAP and DNA template along with any additional enzymes in an artificial organelle enclosed in a cation-specific-permeable exosome;
would such an artificial organelle require its own energy source in the form of mitochondria
how about manufacturing these artificial organelles externally and then delivering the pre-packaged readout machines to selected cellular targets;
how would we go about positioning and anchoring such pre-packaged machines in appropriate locations such as an axon hillock or synaptic terminal;
what about simple expedients for readout like flushing the written tapes from the CNS and routing them to the kidneys for elimination and subsequent external sequencing;
what about directly reading off the nucleotides as the DNAP does its job, transmitting an appropriate code using floating RF transmitters or fluorescent indicators and external imaging to complete the readout; ignoring the problem of accomplishing this on a large scale, what are the main obstacles to doing it at a smaller scale; this option has the potential advantage of allowing the recording apparatus to continuously recycle a supply of nucleotides.
April 25, 2013
Yesterday we heard from Stephen Smith on synaptic diversity, biomarkers of synaptic activity, finding the engrams of experience in the brain, having prepared for his visit by listening to his previously recorded lecture at the University of Colorado in 2012. Here are a few points that were discussed in class — the MM:SS prefix indicates the offset of the relevant segment in minutes and seconds from the beginning of the video found here:
14:11 — diversity of function, regional restriction of mRNAs, regional restriction of protein isoforms, subcellular restriction of proteins — it would seem that the prospects for simulations that don’t make use proteomic and genomic signatures are poor. The opportunities for combinatorics are staggering — do you really think that there are that many qualitatively different functions? It seems to imply that we would have to learn models for all these neuron types and their respective distributions of synapses in order to build simulations of the sort that Henry Markram has been developing and even for a single cortical column the diversity in the 60,000 or so cells that comprise a cortical may considerably complicate the EPFL / HBP project.
However, when asked about Markram’s project, Steven’s primary complaint was that even if we were to learn to simulate a cortical column, it wouldn’t be interesting — or useful — without incorporating additional environmental information including the concentrations of diffuse neuromodulators such as dopamine, serotonin, acetylcholine and histamine and the dynamic and rapidly evolving distributions of ion channels on neural processes.
19:50 — the idea of a connectomic code that tells the pre-synaptic and post-synaptic neurons how they’re supposed to behave, their potential for plasticity and hints about recent short-term memory, or possibly encoding the age and stability of connections implicated in long term memory. Relative to our discussions with Bell and Olshausen last week it might be interesting to investigate whether we could infer proteomic signatures from spiking data or visa versa.
23:10 — array tomography and beyond; four antigens per cycle and 10 to 16 cycles per slice yielding as many as 60 total proteins imaged, with more antigens and considerably higher resolution than is possible with confocal microscopy in which resolution is dependent on the depth of the tissue.
35:20 — training an SVM to identify synapses based on hand-labeled; synapse distributions as a function of cortical depth. Also looking at unsupervised clustering of synapse types. Stephen invited the students present to drop by his lab and check out all the expensive toys and was particularly interested in computer science students with machine learning expertise to collaborate with his team on learning unique synaptic signatures and automatic labeling of synapses.
April 23, 2013
Here is the link to the lecture at the McGovern Institute for Brain Research entitled “Experimenting with MEG: Current Trends and Future Prospects” that I mentioned in class. Concerning the question of spatial and temporal resolution for MEG and competing technologies, here is a synopsis: MEG — 1 ms temporal, 1 cm spatial; EEG — 1 ms temporal, 5 cm spatial; MRI — 1 s temporal, 5 mm spatial. Apropos of Jon’s anecdote about Francis Crick, here is an article from Salk Institute’s Inside Salk magazine entitled “Serendipity and Science” that talks about the collaboration between Ed Callaway and the virologist John Young that led to the rabies virus method for circuit tracing. Here is the quote by Crick from the 1979 Scientific American article [1] that Jon read for us:
Sometimes stating a requirement clearly is halfway to seeing how to implement it. For example, a method that would make it possible to inject one neuron with a substance that would then clearly stain all the neurons connected to it, and no others, would be invaluable. So would a method by which all neurons of just one type could be inactivated, leaving the others more or less unaltered.
Please make use of the discussion notes as they include not just summaries of our discussions in class, but nearly six months of additional notes providing short tutorials and summaries of discussions with diverse technology experts collected in preparation for this class. For example, try searching for “terahertz” or “ultrasound” and you’ll find useful references and descriptions of the applications.
I mentioned that next Wednesday’s speaker, Yael Maguire, wrote his thesis14 on antenna15 design for NMR. He developed what was up until recently the most sensitive planar detectors possible for nanolitre volumes. Yael also started an RFID company called ThingMagic focusing on long-range — not near-field — RFID that developed a chip capable of far-field transmission up to 20 meters and could be manufactured for about one cent per unit. The current use-cases driving RFID technology16 don’t align particularly well with the readout problem but the technology is evolving quickly and bears watching. Here is a press release about Yael from the ThingMagic website.
As for a preview of what Yael will be talking about, consider the problem of trying to determine what portion of the brain could be addressed using a single IR illumination source. Start by drilling a hole a few microns in diameter in the skull and inserting a tiny fiber optic cable. Yael’s focus has been on using near-infrared light to selectively power up circuits implemented as really small, really low-power devices. He showed us the area of cortex that could be activated as predicted by his simulations assuming 350, 35 and 3.5 nanowatts per device.
The problem of controlling a particular neuron seems conceptually straightforward. Send in a coded light sequence that would activate a reporter protein on a single target neuron based on a specific time-code mapping and then use a global signal — a single pulse — into the brain so that only that neuron would fire. In principle, you could do something similar on the readout side with an appropriate readout protein. The challenge is that the light produced by the readout protein is likely to be incredibly weak and so getting the signal out through all the intervening tissue is a daunting challenge.
Apropos of controlling neurons, Oleg found this interesting paper by Nikolenko et al [118] using two-photon illumination. Note that uncaging glutamate is the “other” popular method of photostimulation:
Photostimulation methods fall into two general categories: one set of methods uses light to uncage a compound that then becomes biochemically active, binding to a downstream effector. For example, uncaging glutamate is useful for finding excitatory connections between neurons, since the uncaged glutamate mimics the natural synaptic activity of one neuron impinging upon another. The other major photostimulation method is the use of light to activate a light-sensitive protein such as rhodopsin, which can then excite the cell expressing the opsin. Source: WikipediaThe paper is available here and I’ve included the abstract below for your convenience:
We introduce an optical method to stimulate individual neurons in brain slices in any arbitrary spatiotemporal pattern, using two-photon uncaging of MNI-glutamate with beam multiplexing. This method has single-cell and three-dimensional precision. By sequentially stimulating up to a thousand potential presynaptic neurons, we generated detailed functional maps of inputs to a cell. We combined this approach with two-photon calcium imaging in an all-optical method to image and manipulate circuit activity. Source: [118]
April 21, 2013
For our present purposes, a neural circuit consists of a set of neurons and axodentritic synapses. Define the periphery of the circuit to be those neurons whose axons do not connect to any other neurons in the circuit. Tracing a circuit consists of identifying the neurons and synapses comprising the circuit within a sample tissue and marking them in some fashion, say by engineering the cells to express a fluorescent dye and then imaging the expressed fluorophores.
The research from Ed Callaway’s lab that we discussed in Monday’s class offers one promising technology for tracing circuits. This technology depends on the use of a family of retroviruses that allows us to transfer information in the form of viral RNA by utilizing existing cellular machinery for fast retrograde transport, in which various molecules involved in cellular processes are routinely conveyed from axon terminals to the soma.
Fast retrograde transport returns used synaptic vesicles and other materials to the soma and informs the soma of conditions at the axon terminals. Some pathogens exploit this process to invade the nervous system17. They enter the distal tips on an axon and travel to the soma by retrograde transport. Examples include tetanus toxin and the herpes simplex, rabies, and polio viruses. In such infections, the delay between infection and the onset of symptoms corresponds to the time needed for the pathogens to reach the somas. Source: Wikipedia
We are looking at Ed Callaway’s work primarily for the technology that was developed in his lab for tracing neural circuits using a strain of G-deleted rabies virus (RV) where the G stands for glygoprotein, also known as an envelope protein. This envelope protein plays a key role in spreading infection by coating the viral ribonucleoprotein — see Figure 1 — so that the virus can spread across a synaptic cleft, backwards as it were, from the post- to the pre-synaptic neuron. He also makes use of recombinant DNA technology to modify the host cells (neurons) in order to control propagation (transsynaptic spread of the infection) from one neuron to its synaptically-connected neighbors.
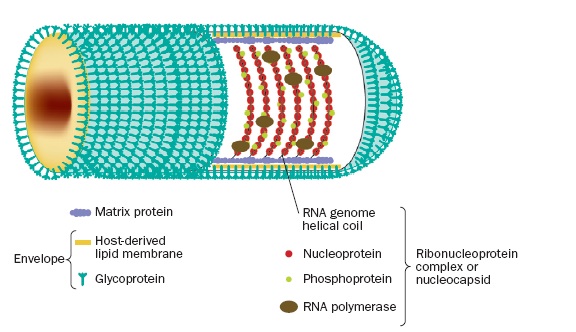 |
The machinery for circuit tracing was developed over several years and published in a series of papers [30, 105]. The first step of creating a strain of virus that doesn’t propagate was accomplished by deleting the rabies viral coat glycoprotein from its genome and replacing it with enhanced green fluorescent protein (EGFP) which is used to image the circuit. Since the glycoprotein is necessary for infection — see Figure 2, its deletion results in an inert virus. However, when raised in complementing cells that express the glycoprotein, the rabies incorporates it into its coat, resulting in an active virus.
 |
Mammals don’t produce the glycoprotein, and therefore the virus can only infect individual cells and will not spread. Since the virus infects through axon terminals however, you can use it to trace inputs to a specific area by injecting it into the area of interest. That is to say, you inject the modified RV into the axons of the neurons you want to trace, and the viral RNA is transported back (in the retrograde direction) along the axon to the cell body where EGFP is expressed so that when the tissue is illuminated by an external light source the cell bodies corresponding to the infected axons “light up”.
The resulting technology is limited to tracing the simplest circuit, a single “wire” from its terminus (synapse) to its origin (soma). To trace more complicated circuits, we need to selectively infect one or more neurons with an RV capable of transsynaptic spreading — typically neurons in the periphery of the target circuit, and then engineer the rest of the cells to express the marker dye (EGFP) but not allow infection to spread further. These steps are implemented with further changes to the virus genome and new lines of transgenic mice that express additional proteins which, together with the new viral proteins, implement the desired programming.
Ed’s talk and the above-referenced papers provide the details of the genetic engineering. However, as an exercise, you might try to imagine how this machinery would work — give your creative side a chance to figure out a (possibly new) mechanism before becoming biased by hearing how Callaway and his colleagues did it. As a warmup exercise, imagine if you could create a line of transgenic mice engineered so that selected cell types, e.g., layer IV basket cells in primary visual cortex with inhibitory and excitatory inputs, conditionally express a particular protein, e.g., an envelope glycoprotein, only in the presence of a promoter supplied by a modified rabies virus.
Transfection is the process of deliberately introducing nucleic acids into cells. So far we’ve talked primarily about transduction in which the transfer is mediated by a virus, but in the latter part of the talk, Callaway talks about using a second approach called electroporation that mediates transfer by modifying the electrical conductivity and permeability of the cell plasma membrane using an externally applied electrical field. Another technique used in the more targeted circuit tracing methods described in the last part of the talk involves a second “helper” virus applied in an initial phase to prepare the genome for a second round of virally-mediated alterations.
If you want to pursue this technology, search for related papers to learn about the tradeoffs of using current methods. In addition to the work mentioned in Callaway’s lecture, follow the graduate students who did the lab work, e.g., Ian Wickersham, and see how they’ve extended the technology as they’ve moved on postdoctorally, e.g., Ian is now a Research Scientist in Ed Boyden’s Synthetic Neurobiology Group at MIT. This posting by the otherwise anonymous Naïve Observer provides a reasonably accurate summary of the innovations discussed in Callaway’s lecture. Combined with Callaway’s papers, this “Cliffs Notes” summary provides a good start for where to look next.
April 19, 2013
I spent most of Friday at Berkeley giving a lecture and talking with scientists and students at the Helen Wills Institute for Neuroscience and Redwood Center for Computational Neuroscience, both at Berkeley. After my lecture, I talked with several highly-respected theoretical neuroscientsts including Bruno Olshausen (director of Redwood) and Tony Bell. Bruno collaborated with David Field at Cornell to develop a model of sparse coding that resulted in one of the most important computational neuroscience papers of the last decade [120]. Tony, while at the Salk Institute working with Terry Sejnowski, developed a highly-influential learning algorithm [14] called Independent Components Analysis that has been used to explain learning in the visual and auditory cortex [154, 155]. I mention this because they belong to a small cadre of scientists working to develop mathematical theories to explain the sort of neural activity BAM/BRAIN is meant to address.
As the initiative has been framed by scientists such as Paul Alivisatos and George Church — a framing that has so far survived bureaucratic efforts to recast its objectives to make them politically more palatable, BAM/BRAIN is all about technologies that will allows us to peer deeper and more comprehensively into the brain. The amounts of data are truly staggering and hardly anyone in the current crop of neuroscientsts have the background to appreciate, much less — and here’s the rub — how they’ll manage the storage or carry out the necessary computation-intensive analysis.
By way of context, the society for neuroscience has about 40,000 members world wide, it publishes The Journal of Neuroscience which is arguably the premier venue for neuroscience papers, and in recent years attendance at the annual meeting has topped 30,000. Among the population of neuroscientists, a vanishingly small number understand anything of computation, what it means generally and what it might mean in understanding the brain — they just don’t have the training and typically harbor crippling misconceptions. It’s hard to convey the depth of this ignorance or the potential consequences for making progress. In particular, it means that there will be little encouragement or contribution from this quarter in figuring out how to scale neuroscience to take advantage of the exponentially increasing returns from computation.
In my discussions with scientists who should be the ones to benefit from and contribute to the aspirations of BAM/BRAIN over the last six months or so, I have tried to ease them into scenarios of the sort that BAM/BRAIN technologies would enable. For example, what if you could sample ion concentrations in a block of neural tissue — let’s say a volume 1 mm on a side — of your choosing from an awake behaving brain at 10 nm spatial resolution and 1 ms temporal resolution? For one thing you would deluged with data: 100 10 nm strides per μm, 1000 μm per mm, 1000 samples per second. Assume 8 bytes per sample rounded up to 10 to keep things simple, then we have 100,0003 × 1000 × 10 or 1019, which is 10,000 Petabytes or 10 million Terabyte disk drives. And that’s for just one second!
But let’s assume for the sake of argument, that we have the storage and computing resources to analyze such gargantuan datasets. Well, we might be able to test some existing theories, for example those of Marcus Meister [82] pertaining to direction-sensitive neurons in retina or perhaps gain some understanding of what all the horizontal axons are doing in the first layer of cortex and whether the various tangles of dendro-dendritic connections found about the primate brain are playing an important computational role — it would seem that they must be but we really haven’t a clue what.
At lunch, Tony Bell joked that BAM technologies might enable us to build a version of the “cerebrum communicator” featured in the short clip from The President’s Analyst that I played at the close of my talk. Tony spun a gedanken experiment in which the corpus collosum is severed and one hemisphere of the brain is replaced by its counterpart from another brain. We discussed how the two hemispheres might sort out the connections to enable communication of sorts. Then we considered instrumenting the corpos collosum with optogenetics so the left hemsphere could be silenced and the data collected from someone else’s brain and fed into the right hemisphere to support brain-to-brain “phone calls”.
When it came to making sense of the deluge of data resulting from recording from our 1 mm cube of neural tissue, Tony was more skeptical. We talked about trying to eliminate hidden variables by recording “everything”, the complications of more diffuse forms of signalling, the distribution of ion channels along axons changing in the space of time needed for the propagation of a single action potential.
As for therapies to treat brain disorders, diseases like Parkinson’s, Alzheimer’s, ALS and depression are unlikely to benefit from the more exotic types of BAM/BRAIN technology, since more conventional pharmacological studies are more effective and expeditious in developing treatment. If we were able to sample more widely, say, neurons in several cortical areas simultaneously, we might gain some insight into schizophrenia but methods for sifting through the deluge of data produced in the scenarios I’ve been suggesting are in their infancy and the computational power that exists today for their analysis is woefully inadequate.
This isn’t to say BAM/BRAIN technologies will be irrelevant to clinical practice. Three of the most interesting recent papers coming out of Karl Deisseroth’s lab address the prospects for direct optogenetic control and feedback to implement interventions in cortical circuits for clinical purposes: the optogenetic modulation of dopamine receptors in the treatment of depression [150], diverse dysfunctional modes including depression and the role of the prefrontal cortex [164] and optogenetic control of the thalamus to interrupt seizures due to cortical injury [123]. All three of these studies would benefit from the ability to record from selected ensembles of neurons to assess the role of short- and long-range connections in the relevant pathologies18.
The problem of inferring function or learning simulation models from input/output behavior becomes considerably easier [9, 42] if you can intervene to activate and silence neurons in the periphery of the tissue block by leveraging optogenetic technologies, but while theoretically more tractable, practically speaking even with access to most powerful supercomputers at Livermore, Oak Ridge and Los Alamos the analysis alone would take years and we know from experience that unless you can iterate quickly the prospects for progress are dim.
Olshausen, Bell, Mike Hawrylycz at the Allen Institute and other computational-savvy scientists are generally pretty optimistic about the prospects for computing, and they know how much the next Intel tick-tock cycle can make a difference in our ability to understand complex dynamic and emergent phenomena like the brain, weather, climate, etc. They are also well aware of the fact that the lion’s share of $100M will go to DoD, DoE, NIH with only a small fraction going to NSF to be overseen by Obama’s dream team of Cori Bargmann and Bill Newsome. For the DoD, the technological advances associated with BAM/BRAIN are much more closely aligned with their interests.
BCI (Brain Computer Interface) technologies would benefit enormously from BAM technologies. However a “mindmeld for the warfighter” is likely to become feasible not because BAM/BRAIN will yield insight into the function of neural circuits, but because when you wire an appropriate chunk of neural tissue to some relatively-straightforward signal-processing machinery attached to, say, a remote-controlled drone, the brain is amazingly effective at sorting out the signals and adapting them to directly control the hardware, bypassing much slower control paths [11]. Not surprisingly, advanced weapons, immersive gaming and interactive entertainment will be the big winners short term.
I have to say that Bell, Olshausen and the packed audience that turned out for my talk were excited about the potential technologies that might surface from BAM/BRAIN. The prospects for such technologies arriving on the scene could result in a sea change in the way we educate the next generation of neuroscientists, as well as introduce innovations that are both world changing and frightening to many in their implications for society.
It’s my opinion that many of the health benefits used to justify the cost BAM/BRAIN are more likely to come from other avenues as there is money to be made and more expeditious means of exploiting low-hanging fruit. Moreover, initial funding will primarily benefit entrenched interests and serve to maintain the status quo, input from Bargmann and Newsome not withstanding. Practically speaking however, the BAM/BRAIN initiative will help to re-set the bar and the longer-term benefits of the technologies it seeks to foster will be enormous.
April 17, 2013
After class, we briefly discussed the idea of writing a jointly-authored technical report surveying BAM-related technologies and evaluating their potential value both in terms of the grand-challenge aspirations of BAM/BRAIN and in term of the near- and longer-term prospects for making them available to scientists for their research. The idea is that we restrict our attention to one or two recording technologies, e.g., in the introduction I suggested calcium imaging in the axon terminal and voltage thresholds in the axon hillock.
In order to keep the overall report manageable — both in terms of our writing it and others reading it — we will focus almost exclusively on reporting — or “readout” — technology. Unless the target reporting technology is incompatible with our focus recording technologies or there is an alternative recording technology that works particularly well with the target reporting technology19 we will adhere to this discipline.
Here are some of my notes in trying develop a simple, easy-to-follow structure for organizing the technical report:
Characterization of the two basic components comprising every “complete” BAM technology:
Recorder — think of a recording device that when you hit the record button converts energy from the microphone (sensor) into a semi-permanent record (physical encoding) of the signal for subsequent reuse; the notion of recorder combines the functions of sensing the target signal and encoding it a form suitable for transfer.
Reporter — think of a news reporter who seldom observes the actual events but rather collects first-hand accounts, writes her interpretation of events and posts them to her editor in a remote office; the notion of reporter combines the functions of transcoding, perhaps compressing and transmitting recorded information.
Recording Technology:
Fluorescent — signal by photon emission from natural or inorganic fluorophores coupled with biological sensors that sense voltage, proteins, etc;
Magnetic — convert cellular information into a pattern encoded in magnetic domains;
Electrical — signal local electric potential by exploiting how fluctuations in ion concentration effect the DNA polymerase replication error rate;
Genetic — signal by converting cellular information into DNA or RNA sequences using existing biomolecular machines or nanoscale versions of gene sequencing technology;
Mechanical — signal encoded in the deformation of protein complexes or microbubbles;
Reporting Technology:
Imaging — using an external energy source radiating in the electromagnetic or acoustic spectrum to illuminate reporter targets and then collecting reflected energy to read out the signal2021
Local Transmission — reporters are coupled to local transducers that convert one or more signals to a format suitable for transmission over a communication channel targeting an external receiver2223
Here are a couple of questions that were asked at the end of Kevin Briggman’s talk along with Kevin’s answers:
TLD: How do you think CLARITY will change your research or that of other researchers at NIH or elsewhere with whom you collaborate?KLB: We will use it, of course, for protein distributions in brains. For that I think it will be great. For circuit tracing, I don’t see how you can use it with light microscopy. If you had everything labeled, the wires are going to be very difficult to resolve. For example, if every axon in the zebrafish were labeled with a fluorescent protein, you wouldn’t be able to resolve them using light microscopy. I think it’s a great technology if you have sparse connections. We’ll probably use it in conjunction with antibody labeling to get protein distributions. However, in terms of trying to follow every wire, I think you’re still going to be limited by diffraction in light microscopy.
TLD: Does the fact that the cellular scaffolding is replaced with a hydrogel and the lipids are washed out make it more difficult to trace axons and dendritic branches?
KLB: I think so. I didn’t mention this in my talk, but most of what our heavy metals label is lipid. Lipid is what defines the cell membrane. So, yes, eliminating the lipid is counterproductive if you’re trying to distinguish the border between cells. One of the greatest ways of tracing a neuron is to label it with a lipophilic dye. These are classical dyes that diffuse into lipids and become extremely bright, so you can basically follow a halo of light as you move along an axon, and those dyes would not be compatible with CLARITY.
TLD: Would you still use the serial-section block-face SEM or would some form of two-photon, deep-penetration imaging suffice?
KLB: Even if the index of refraction is well matched to two-photon imaging, there is an ultimate z-limit due to the residual scattering from proteins in your sample. I think it just comes down to an issue of density. If all 100B neurons in a brain are labeled, you’re not going to be able to see very far in a CLARITY-prepared brain. If the labeling is sparse, then distinguishing individual neurites might be done without sectioning at all.
BOA: Is there a fundamental limit to how fast you can scan?
KLB: The fundamental limit is determined by the physical properties of our electron detector. We use a silicon photodiode, and that diode has a capacitance and resistance associated with it which define a time constant for the photodiode limiting the slew rate of current in the photodiode so you can’t exceed that time constant. You can try to reduce the capacitance or resistance of the diode but only so much and we’re pretty much limited by a maximum 10MHz slew rate.
KLB: There’s a new approach advertised by Jeff Lichtman and others that involves high-throughput, large-area imaging using a new multi-beam scanning electron microscopy technology just introduced by Carl Zeiss. It splits the electron source into multiple beams — as many as sixty – and then scans and records all sixty in parallel. So that gets you a 60-fold speedup in scanning. If this works — it uses an entirely new method of detection which we have yet to evaluate — we’d get a 60-fold speedup in acquisition rate. That’s obviously huge. Even so, a mouse brain is daunting given that the bottleneck is now analysis. It would still take on the order of ten years if you crunch the numbers, but it makes it doable within our lifetime. We’d need a 60-million-fold speed up in our current methods of analysis to keep up with the new acquisition rate.
TLD: Kevin mentioned light-sheet microscopy as a method that may make it easier to carry our accurate histological reconstructions. The Ahrens et al [4] paper from the HHMI Janelia Farm Laboratory uses light-sheet microscopy for whole-brain imaging in zebrafish embryos.
KLB: Regarding circuit tracing with CLARITY, I should have added that the problem of tracing (even multicolor) densely labeled axonsmand dendrites with diffraction-limited optics is basically the same problem with using Brainbow for tracing. Despite generating compelling multi-colored images, when one zooms in on the neuropil of a densely “Brainbow-ed” animal, individual axons and dendrites are unresolvable. I’m fairly certain even Jeff Lichtman would agree with this assessment.
All of these statements of course come with the caveat of using diffraction-limited optics. I think there is considerable room for improvement in methods for large-scale super-resolution 3D optical techniques that may make many of the labeling techniques — CLARITY, Brainbow, dense virus labeling, etc. — useful for circuit tracing by breaking the diffraction limit. A number of labs, including my own, are working on scaling up such techniques.
In an earlier post, David Cox wrote:
The biggest challenge that I see with all of these endeavors has less to do with the number of neurons (which is what BAM seems to be primarily concerned with) and more to do with how much data one can get from each neuron (which BAM may very well address, although it is not clear what BAM is at this point).David Heckerman’s replied:
I agree and am reminded about a project I did in med school in the early 80s, where I surveyed the information processing capabilities of neuron branching. It turns out that the exact nature of the branch (e.g., diameters, cross-sectional shapes, branching angles) can greatly influence which pulses travel down which branches, making the branch a first-class information processor.I plan to follow up on Heckerman’s mention of branches as active computing units — just one of many ways in which computing is distrbuted with in the cell body, but I want to comment on the issue of recording from many versus few neurons simultaneously. One take-away message from Briggman’s observations regarding CLARITY is that cluttering up the target tissue with recording proteins increases noise due to light scattering and so sparsity is key in operations that require, for example, resolving axons and dendrites. This suggests that traditional imaging techniques that rely on wholesale illumination may not scale to millions of densely packed neurons.
April 15, 2013
Following a suggestion from Jon Shlens, I tracked down this paper [166] reporting on the development of a line of transgenic zebrafish that are transparent throughout their lives. The study from Janelia Farm that came out a couple of weeks ago [4] was based on zebrafish in their larval stage.
Zebrafish are genetically similar to humans and are good models for human biology and disease. Now, researchers at Children’s Hospital Boston have created a zebrafish that is transparent throughout its life. The new fish allows scientists to directly view its internal organs, and observe processes like tumor metastasis and blood production after bone-marrow transplant in a living organism. (source)Thinking about these recent papers that exploit various means of rendering neural tissue transparent is a good approach for exploring possible project proposals and formulating questions for our visitors about how these new methods and findings impact their thinking and directions for future research.
With some help from David Cox, I learned a little more about zebrafish, their characteristics pertinent to optical transparency and our ability to image their internal structure. The quick summary is that it is only the skin that is transparent in larval zebrafish, subdermal structures with chromophores that produce significant light scattering still limit effective penetration depth. However, by avoiding scattering in the normally pigmented superficial layers and owing to the small size of the larval organism, the developing brain is small enough that it is well within the feasible recording depth for 2-photon imaging — which is a useful technique, in part, because it is reasonably tolerant of scattering of collected photons. In general, blood, melanin in the skin, fat and water — in the case of the frequencies (800 nm to 2500 nm) used in near-infrared spectroscopy — are the tissue components most responsible for absorption.
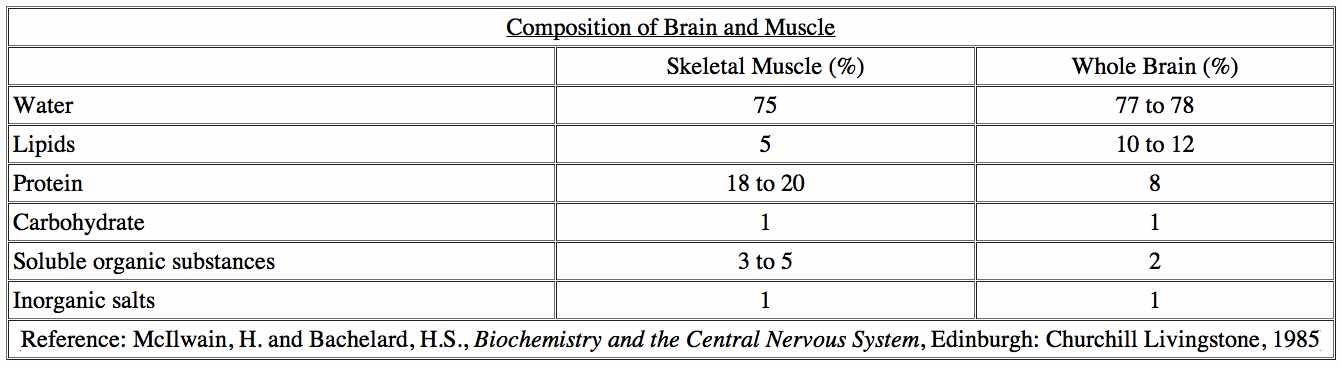 |
In mouse and human systems, the in vivo spatial resolution of the adult animal is limited due to the normal opacification of skin and subdermal structures. The characteristic adult pigmentation pattern of the zebrafish consists of three distinct classes of pigment cells arranged in stripes: black melanophores, reflective iridophores, and yellow xanthophores. Some mutant strains exhibit a complete lack of one or more of these types of pigmentation (source). White et al [166] developed a transgenic strain of zebrafish that largely eliminates all three in adult fish, but does nothing to reduce absorption in subdermal structures.
April 13, 2013
Kevin Briggman will be talking on Monday. There are two papers [25, 24] on the calendar page and you should at least skim their abstracts and introductions prior to class. His slides are also accessible on the calendar page and you might want to scan them to help formulate questions for class discussion. Kevin will be talking about the technology for serial block-face scanning electron microscopy (SBFSEM) which is used in connectomics and other neural imaging applications. He is broadly knowledgeable in neuroscience and the relevant physics of electron microscopy and so you should feel free to ask him questions about related technologies.
Mike Hawrylycz will be talking on Wednesday. Two papers [144, 65], his slides and a video lecture are available on the web site. Watch the lecture, take notes and formulate questions for class. Mike knows a great deal about the technologies used by the Allen Institute for the Mouse Brain Atlas and is working to develop the infrastructure for an atlas of the human brain. The mouse atlas relied on a wide range of imaging technologies for connectomics and proteomics. One of the biggest challenges facing the human atlas involves standardizing across the brains of multiple subjects. Hawrylycz et al [65] describes how the Allen Institute team addressed this problem in mice — a considerably simpler problem but challenging nonetheless.
For your own benefit, spend a couple of hours prior to class working through the materials that we’ve made available on the course website. Your preparation will be paid back many fold in terms of what you’ll get out of the in-class discussions and by being better prepared to take advantage of the opportunity to ask our invited speakers questions about their areas of expertise as well as questions that could help you flesh out your project proposals.
April 11, 2013
Ed’s lectures are linked off the course calendar page. You can also access them here. I’ve also added a bunch of slides from Ed’s “Principles of Neuroengineering” course at MIT that include most of the slides he showed us Monday and Wednesday, plus some additional material that fleshes out the trajectory — from macro to micro scale — outlined in his talk. In anticipation of discussions involving fluorophores, quantum dots and FRET which stands for either “Förster Resonance Energy Transfer” or “Fluorescence Resonance Energy Transfer”, I prepared a short introduction which you can find here.
After class on Wednesday, Dan asked about the evolution of the prefrontal cortex (PFC), the idea that the difference between apes and humans might be merely one of the size and complexity of cortex, and whether the advanced language, social and abstract reasoning capacity of humans could be accounted for by differences in cortical structure. This paper [43] (PDF) published last year directly addresses those questions. There’s also a public lecture (VIDEO) (SLIDES) if you’re interested.
A new tissue preparation technique out of Karl Deisseroth’s lab renders an entire mouse brain essentially transparent [35]. Moreover, the process “preserves the biochemistry of the brain so well that researchers can test it over and over again with chemicals that highlight specific structures within a brain and provide clues to its past activity.” One potential disadvantage of the technique is that it washes out the lipids. The technique makes use of a hydrogel which “forms a kind of mesh that permeates the brain and connects to most of the molecules, but not to the lipids, which include fats and some other substances. The brain is then put in a soapy solution and an electric current is applied, which drives the solution through the brain, washing out the lipids.”
I had assumed that washing out the lipids would make it difficult if not impossible to resolve cell boundaries but the authors report that using mouse brains “we show intact-tissue imaging of long-range projections, local circuit wiring, cellular relationships, subcellular structures, protein complexes, nucleic acids and neurotransmitters.” Moreover their preparation also “enables intact-tissue in situ hybridization, immunohistochemistry with multiple rounds of staining and de-staining in non-sectioned tissue, and antibody labelling throughout the intact adult mouse brain.” Pretty amazing technology. Of course, there’s no chance of recording any dynamics, but still it could be a real game changer for the field of connectomics.
The HHMI Janelia Farm work on imaging transparent fish does offer opportunities for observing the dynamnics of neural activity. You might want to take a look at the Ahrens et al paper [4] on whole-brain, cellular-level imaging of the zebrafish and ask yourselves what, if anything, you — or a well-crafted machine-learning algorithm — might be able to infer from analyzing the data. See the news article and video for inspiration. By the way, the Chung et al [35] method works by washing out lipids, but the zebrafish must have transparent cell membranes which presumably are made of the same bi-lipid layers that our cell membranes are made out of since it seems implausible that zebrafish use an entirely different chemistry to create their cell membranes — see here for a follow-up discussion relating to this issue.
April 9, 2013
The audio from yesterday’s discussion with Ed Boyden is now up on the course calendar page. We had some technical problems with the audio that we expect to have fixed by tomorrow. The audio problems resulted in our losing the first few minutes of the lecture, and so I added a short preface to provide some context for the point in the discussion where we join Ed. You might want to listen to the lecture again and think about any questions that you’d like Ed to answer during tomorrow’s lectures. If you track down any references that Ed mentioned while you’re listening to the recording, please send the links to me and I’ll post them on the discussion page.
The lost portion of the audio ended just at the point Ed was talking about a recent paper [64] in Cell that suggests glial cells play a more important role in human cognition than previously thought (VIDEO). The authors of the paper in Cell first isolated glial progenitor-cells in the central nervous system, these cells enable the growth of astrocytes — a type of glial cell —- from human brain tissue. These cells were then transplanted into the brains of newborn mice. As the mice grew, the number human glial cells exceeded the number mice glial cells, but left the existing neural network of the mice intact. The researchers found that two important indicators of brain function had improved dramatically in the mice with the transplanted human glial cells.
First, they considered the speed at which the signal is transmitted between adjacent astrocytes in the brain, and they noted that the transmission speed in mice with transplanted human glial cells is faster than in normal mice, similar to the transmission rate measured in the tissues of the human brain. And second that the mice with transplanted human glial cells developed faster and more sustained long-term potentiation, which means that the learning ability of these mice increased. The researchers also ran tests to measure memory and learning ability. They found that mice with transplanted human glial cells learned more quickly, they gained new associations faster, and they performed a number of different tasks significantly faster than mice without the transplanted human glial cells.
This morning I was talking with a former student of mine, now neuroscientist, Arjun Bansal, who is doing a postdoc in Gabriel Kreiman’s lab at Harvard. Arjun was telling me about a theoretically possible signalling pathway in bacteria that might be relevant to whole-brain recording. The basic idea is outlined in this article in MIT Technology Review magazine. The article begins by describing a controversial hypothesis that bacteria can transmit radio waves. It than goes on to report that physicists now claim to know how bacteria could accomplish this feat, at least theoretically. As Ed was saying yesterday, if there’s a physical way to do something — in this case signal wirelessly using radio waves, then it is likely nature has implemented it multiple times over the millennia.
Chemical reactions can be induced at a distance due to the propagation of electromagnetic signals during intermediate chemical stages. Although it is well known at optical frequencies, e.g. photosynthetic reactions, electromagnetic signals hold true for much lower frequencies. In E. coli bacteria such electromagnetic signals can be generated by electrical transitions between energy levels describing electrons moving around DNA loops. The electromagnetic signals between different bacteria within a community is a “wireless” version of intercellular communication found in bacterial communities connected by “nanowires”. The wireless broadcasts can in principle be of both the AM and FM variety due to the magnetic flux periodicity in electron energy spectra in bacterial DNA orbital motions. Source: Electromagnetic Signals from Bacterial DNA, by A. Widom, J. Swain, Y.N. Srivastava, S. Sivasubramanian (HTML)
April 7, 2013
I contacted a number of people about the question of fitting models to high-fidelity, high-resolution cellular data. My poll of experts included: David Cox, David Heeger, Geoff Hinton, Ray Kurzweil, Jerry Pine, Marc’Aurelio Ranzato, Eero Simoncelli, Mike Tarr and Daniel Waganeer. Here is a sampling of what I learned either directly from them or by perusing the websites of other scientists they pointed me to.
Jerry Pine’s lab develops micro-fabricated neuron cages that can be laid out in an array with one neuron placed in the center of each cage to attach and grow processes out through tunnels in the cages. An electrode in the center of each cage provides for long term non-destructive two-way communication with the cell, to stimulate it or record from it. Source: Erickson, J. C., Tooker, A., Tai, Y-C., and Pine, J. (2008). Caged neuron MEA: A system for long-term investigation of cultured neural network connectivity. Journal Neuroscience Methods 175:1-16.
Daniel Wagenaar’s lab tackles the problem of studying how information flows through the nervous system by recording from a large number of cells at once. They propose to meet this challenge by “using voltage-sensitive dye imaging and multi-electrode array (MEA) recording. The combination of these techniques will make it possible to record neural activity with single-cellular spatial resolution and single-action-potential temporal resolution.” Wagenaar is also known for his early work on neurally controlled animats. Source: T.B. DeMarse, D.A. Wagenaar, A.W. Blau, and S.M. Potter, 2001. The neurally controlled animat: Biological brains acting with simulated bodies. Autonomous Robotics 11(3), 305-310.
Geoff mentioned work by David Heeger, Tony Movshon and Eero Simoncelli. David replied to my inquiry suggesting that I “have the students first implement a conventional model of some kind, use that to generate simulated data, and then feed the simulated data (because you can generate as much as you want), into a feedforward neural net. Attached is a recent review paper from which you should be able to get enough info to implement such a simulation.” David included a paper by Carandini and Heeger [31] and I found this video of his lecture describing his basic theory of normalization as a canonical neural computation. David’s response wasn’t exactly what I was looking for and I tried to explain further:
Several projects are looking at whole-brain recording at greater temporal and spatial resolution. Others are looking at smaller target circuits but with much higher resolution involving many inter-, intra- and extra-cellular readings. The point of the exercise after class last week was to come up with compelling use cases. Suppose you could determine for each reading whether it originated from inside of a given neuron or outside, along with coordinates for the positions within the cell or in the extracellular matrix. I grant you it’s a little hard imagining how this could be accomplished but that was our starting assumption. The question then is: could you learn an accurate predictive model of the neurons so that you could simulate the observed behavior? How much would it help if you were able to activate or inhibit individual neurons using additional mechanical or optogenetic probes?
David Cox provided the most detailed response, excerpts of which I include below:
A few quick thoughts on your question: There’s a bit of a bias in neuroscience against models that explain or predict the data in the absence of some claim of structural verisimilitude or some neuroscience “lesson” to be learned. Here are some leads on work I know about; happy to provide more if it is helpful:
Tommy Poggio has done a fair bit of work fitting HMAX models to anybody’s-data-he-can-get-his-hands-on (e.g. Charles Cadieu’s work when he was student there (PDF). Jack Gallant at Berkeley has done a fair bit of model fitting for individual unit responses, though, again, not so much for networks per se. As Geoff notes, people like Simoncelli, Heeger and Movshon have also done work fitting models to electrophysiological data e.g. in V1.
Not quite what you’re asking for, but possibly related: Nikolaus Kriegeskorte has done some interesting work in comparing neuronal systems to artificial systems, and comparing multiple data sets to each other (PDF). I just learned that Mike Tarr’s group at CMU is now doing similar work as well (not yet published).
Also not quite what you’re asking for, but there is a whole sub-genre of fitting more biologically-realistic models (a la Hodgkin-Huxley) that your students might be interested in. You might get more traction tapping this literature by adding the search term “biophysical”, or the name of a particular simulator package (e.g. “Genesis” or “NEURON”"). There’s a large collection of such models, many of which target specific neurophysiology data set (HTML). These kinds of modeling efforts often have very different goals from what I think you are after, but it might be useful for your students to look here. Markram sits somewhere on this spectrum, and you can interpret that however you like.
The biggest challenge that I see with all of these endeavors has less to do with the number of neurons (which is what BAM seems to be primarily concerned with) and more to do with how much data one can get from each neuron (which BAM may very well address, although it is not clear what BAM is at this point). Most models have a huge number of free parameters, and it is not obvious that any currently available experimental techniques collect enough data to meaningfully fit such models. For, instance, Tommy can fit many things with an HMAX model if you give it enough free parameters, but typical single-unit physiology experiments that these fits are based on often only involve 20 minutes to an hour worth of data per neuron, so I’m concerned that almost any sufficiently expressive model could fit that data.
You can get more data from an anesthetized monkey (as Tony Movshon does), but this is not suitable for all systems, since the anesthetized brain is clearly not in its ordinary operating range. Neurons in IT cortex, for instance, have greatly altered and suppressed responses in anesthetized animals. Also, the space of stimuli you’d like to span to meaningfully constrain a model is extremely large (as an aside, one of the reasons that we’re pursuing two-photon imaging now is the promise of getting many days to work with the same cells so that we can have a prayer of getting enough data from individual neurons, both to characterize them and to see them change as a function of learning – at least until our imaging windows fail). Hopefully some attention gets paid in BAM not only to how many neurons, but also how data per neuron can be collected, and with what kind of simultaneity of measurement.
Another place where modeling often gets squishy in neuroscience is in comparing a proposed model to alternatives; often there aren’t sufficiently many (or any) appropriate foils to compare against, leaving the paper sort of “single-ended”. A model is shown to fit “well”, but without a clear definition of what “well” means, or whether a simpler model could offer similar explanatory power. However, your goals for this kind of data might be very different.
Thinking more about David Cox’s and David Heeger’s comments, I may have missed an important point relevant to BRAIN. Apropos BRAIN-initiative co-director Bill Newsome’s comments vis a vis recording from smaller samples of cells — now repeated in several news stories about the BRAIN initiative — and independent of whether or not whole-brain recording is possible anytime soon, there is considerable value to be had in recording from smaller ensembles but with much higher temporal and spatial resolution. Moreover there are simulation experiments that we can carry out now, both to test the hypothesis that dense recording will yield useful insights to the tools of machine learning and to evaluate proposals aiming to provide more detail in terms of what we might learn from such recording capabilities. The 3-D probes being developed in Ed Boyden’s lab are relevant to this class of investigation. Here’s a sketch of what students might do to follow through on this proposal:
Create a simulation of a small neural circuit using the best multi-compartment, Hodgkin-Huxley or alternative such as the Fitzhugh-Nagumo model. The more accurately the model accounts for cellular detail in the target circuit the better, but our goal is not to build better models; the goal is to generate data and in a small circuit we can afford to throw a lot of computation at doing the job as well as current science allows. Now “instrument” the model circuit to collect data in accord with some proposed recording technology. Finally, create training and testing datasets and set up an evaluation protocol in which some of the data is withheld and the task for a learned model is to fill-in the missing data. For example, define a sub circuit of a larger neural circuit, identify the “inputs” and “outputs” for this sub circuit, collect data on the sub circuit and train an input-output model, run the simulation substituting the learned input-output model for the sub circuit, and then evaluate the learned model based on its predicted outputs compared with the full simulation “ground truth”.Apart from student projects, there is a lot one could do with such simulation models and the data they can be used generate. Creating these models would serve to sharpen and define BRAIN-initiative objectives, allow researchers working on recording technologies to get a better idea if their ideas are likely to prove useful to the community, and provide computational neuroscientists and the broader community of machine learning researchers the opportunity to test ideas, anticipate and help shape new technologies, and take on the challenges and enjoy the advantages of working with vastly larger datasets. We could even promote competitions in NIPS and ICML similar to the PASCAL Challenges that have spurred so much innovation in the computer vision community.
April 4, 2013
Here are answers to some of the questions posed by students considering taking the course:
The 2013 version of CS379C with this particular focus and collection of participating scientists is not likely to be offered again any time soon. This is partly because it takes a lot of work to organize and requires additional effort on the part of my collaborators whom I prefer not to impose upon frequently. The venue for presentations will depend on the number and quality of the projects. If appropriate, I’ve offered to convene a panel of scientists and engineers to listen to short project pitches and provide feedback to the students.
I will do my best to record at least the audio for the in-class discussions. This will depend, of course, on the speaker consenting to being recorded and agreeing to allow the resulting recordings to be made public — or at least public to the Stanford community. In cases where our visitor has slides and agrees to having them shared, I will make these available as well, but bear in mind that, possibly excepting Ed Boyden’s two lectures next week, slides may be rare since the in-class discussions are meant to be just that, namely free-form discussions of ideas and brainstorming about proposed solutions to neural recording and readout problems.
If you cannot attend a class, you’ll be able to send your questions to my teaching assistant, Bharath Bhat or the visitor’s student host and, if there is time, we’ll ask your questions for you, though I request that you not to abuse this option. I think the sort of free-form discussions this class features will be more valuable to the students than a formal, canned lecture, especially given that nowadays most researchers or the institutions where they speak routinely post video of their presentations to YouTube.
If you end up not registering for a grade, please do register for audit as that way I will have a record of your participation and it will be easier for me to manage student communications, password-protected resources, etc. Also, if you do choose to audit, I would appreciate it if you would agree to serve as an informal consultant for students involved in projects where your particular expertise may have application. This would require at most minimal effort on your part, but could be very helpful to a student trying to puzzle through some problem outside his or her area of expertise.
Yesterday we talked about the Zador and Kording proposals as starting points for projects with the idea of extending them to address unanswered scaling questions associated with their respective readout problems. All of you should use the Berkeley lecture to start generating ideas for your project proposals and thinking about the constitution of teams to cover the relevant technical problems you foresee. Don’t hesitate to contact me if you want to try out an idea for a proposal.
April 3, 2013
In class today, we drilled down into some of the technologies discussed in the assigned on-line lecture. I noted that the lecture was conceptually organized to take the reader along a trajectory in which we start with a few target goals, e.g., recording action potentials and generating the connectome, consider the state of the art, look at increasingly more complicated tools from nanotechnology and scanning electron microscopy and finally return to look more deeply at methods for re-purposing and re-combining existing molecular machines in order to accomplish our initial target goals. The last few slides provide a glimpse of two such approaches: one addressing the problem of generating the connectome and a second the problem of recording action potentials from large ensembles of neurons.
Specifically we talked about the Zador et al [172] proposal for “sequencing the connectom” and the Kording [88] proposal for a “molecular ticker-tape” to record cellular dynamics. Both of these papers are little more than high-level proposals; they are brief, respectively, six and four pages excluding references, but the basic ideas are biologically plausible and there are now several labs working to develop experimental realizations. In the case of ticker-tapes, the Zamft et al [173] provides a good next step looking into some of the fiddly bits24. There is no Zamft et al analog for the sequencing-the-connectome paper, but Tony Zador tells me that his lab has a couple of papers in the final stages and will share them with the class as soon as they are submitted and hopefully before he visits us for class discussion in early May. As an exercise in class, we looked at the three steps in Zador’s “algorithm” for sequencing the connectome: generate unique barcodes, pair the barcodes from connected neurons, and concatenate the barcode-pairs and sequence the resulting DNA. Here’s a recap of what of how to think about the first of these steps:
Either extract a suitably random gene cassette from the cell’s DNA — not likely to be easy — or start with an existing cassette and perform edits to produce a shuffled cassette that is suitably random. Exercise: Do the math: (1) specify the properties of the edit operations, e.g., excise, invert, shuffle, (2) create a model for how these operations are to be applied, e.g., how many times is an operator applied and how are the sites to be selected, and (3) calculate the likelihood that barcodes generated using your method will be unique in a population of a given size. Now drill down and consider available recombinase enzymes and how they might be coerced into generating a barcode. How do you ensure that each cell will generate exactly one barcode? Why is Cre not a good idea? How many instances of this unique barcode will you need? How do you prevent barcode generation from interfering with normal function of the cell? In particular, could your method overtax the cell by depleting its energy budget?
Note that most of the analysis could be done without any additional knowledge about neuroscience beyond basic biology. Zador et al [172] provide hints about choosing enzymes and outlines the basic idea. You can find out about the RCI recombinase that the authors mention in their paper with a quick literature search [61]. You’ll have to make some assumptions about how to control your chosen enzymes, and here you might consult with molecular biologist if you know a convenient source but there are also plenty of papers describing the relevant cellular machinery and I’d be happy to supply a good primer on the requisite biology. The rest of the analysis is a pretty standard exercise in combinatorics, employing the sort of math you learn in an introductory statistics or algorithms course.
There are lots of other loose ends in Zador et al that might be ripe for class projects. How do you apply retroviruses to distribute barcodes to adjoining cells? Ed Callaway might be worth consulting on this. How could you employ ideas from immunochemistry and immunoflorescence25 in particular to assay additional information about cell types? In considering this last challenge think about the problem of deciphering neural circuits and the analogy with Intel’s competition reverse engineering their chips:
Chip companies routinely attempt to reverse engineer the chips of their competitors. Ignoring whether this is legal or not, the standard procedure for doing this provides a nice analogy for how neuroscientists are attempting to reverse engineer neural circults using scanning electron microscopy. Most chips are fabricated in multiple layers applied using photolithography. In order to infer the structure and function of a chip, you have to peel back the layers to reveal the three-dimensional structure of the chip and, in particular, the traces or “wires”, transistors and other electronic components that comprise the circuit. This 3-D reconstruction is accomplished with a slice-and-image SEM approach not unlike we saw in the connectomics work [110, 24, 74].
Inferring silicon circuits from stacks of micrographs is a lot easier than inferring neural circuits26. Engineers pretty much know what transistors look like when etched in silicon. For the most part, they’re also able to recognize logic gates, half-adders, memory registers, flip-flops, etc. The analogous facility for neuroscientists is not nearly so well developed. Detailed neuroanatomical study can yield considerable insight into neural circuit function27 and Zador refers to success in understanding how inhibitory and excitatory connections work in motion-sensing circuits, but such approaches don’t scale well. Perhaps the most widely touted success story concerns our understanding of C. elegans — a common roundworm with exactly 302 neurons and 7,000 synapses give or take a few. In 1996, Sydney Brenner’s lab reported a complete reconstruction of all neural and synaptic connections in this organism. Unfortunately much of what we learned doesn’t translate to human or even mammal brains, since the neurons in C. elegans are non-spiking and behave as complex analog devices with significant functional variability among the 302 neurons.
After class yesterday, we discussed the idea that much of traditional neuroscience research might be supplanted by data-driven model selection using the latest tools and techniques from machine learning. It’s not that we couldn’t have attempted this before, but with new scalable recording technology and the ability to train models with millions of parameters we now have a much better chance of succeeding. Modern (artificial) neural network technology could be employed to utilize diverse data types including not just ion concentrations but also cell characteristics, protein expression levels and voltage potentials.
With enough data and rich-enough models our experience with Google Brain suggests we should be able to learn highly-accurate predictive input-output models for individual neurons and small circuits. As a first test of this idea, we might try to instrument a suitable target organism, e.g., a giant squid axon, so as to record the same sort of data that Hodgkin-Huxley based their early models for the initiation and propagation of action potentials and see if we can obtain an accurate predictive model using an admittedly opaque neural network. I suggested that the students do a literature search for evidence supporting this hypothesis, but I suspect they came up empty handed since I wasn’t able to find anything after spending half an hour’s worth of concentrated effort. In another conversation, we pondered if “generalization-free learning” was an oxymoron or just a misunderstanding of models that are allowed to overfit and yet seemingly extend to unseen exemplars.
April 1, 2013
Tuesday morning, President Obama announced a broad new research initiative, starting with $100 million in 2014, to invent and refine new technologies to understand the human brain. Previously known as BAM for “Brain Activity Mapping” and now called BRAIN for “Brain Research Through Advancing Innovative Neurotechnologies”, the initiative aims to record from and map the “activity of brain circuits in action in an effort to show how millions of brain cells interact.”
This quarter CS379C will investigate the technical feasibility of several recent proposals for addressing these ambitious goals. These proposals include methods for recording action-potential correlates in DNA, reading off the connectome by sequencing and building brain-wide self-organizing nanoscale communication networks. Student teams will evaluate variants of these proposals specifically with respect to the problem of “reading out” neural activity recorded at the cellular level.
Monday’s class provided a general overview and administrative information — see below. The first full class on Wednesday — students are encouraged to read the lecture notes prior to class — will survey several of the most promising technologies relevant to BRAIN. The remainder of the classes will feature scientists from several of the labs involved in the development of this initiative and contributing to key technologies who have agreed to assist teams working on class projects relevant to the BRAIN “readout” problem. Here is the administrative information we went over in Monday’s class:
Administrivia:
Format: Research papers, flipped-classroom lectures and class discussions;
Grading: Class participation plus project proposal, report and presentation;
Prereqs: Good math skills, mastery of basic high-school biology and physics;
Projects: Small teams working to develop solutions to the BAM “readout” problem;
Resources: Tutorials, discussion logs, annotated bibliography, technical consultants:
Kevin Briggman, NIH, (previously at Max Planck in Winfried Denk’s Lab);
Ed Boyden, MIT;
Greg Corrado, Google, (Stanford Neuroscience Ph.D. with Bill Newsome);
David Cox, Harvard;
Mike Hawrylycz, Allen Institute;
David Heckerman, Microsoft, UCLA;
Bruno Madore, Harvard;
Yael Maguire, MIT (Gershenfeld Lab);
Adam Marblestone, Harvard (Church Lab);
Akram Sadek, Caltech (Scherer Lab);
Jon Shlens, Google, (UCSD Neuroscience Ph.D. with E.J. Chichilnisky);
Mark Schnitzer, Stanford;
Ben Schwartz, Google, (Harvard Biophysics Ph.D. with Dr. Nathan J. McDannold);
Stephen Smith, Stanford;
Fritz Sommer, Berkeley, Redwood Institute;
Brian Wandell, Stanford;
Tony Zador, Cold Spring Harbor;
March 31, 2013
In the Berkeley lecture and in my earlier class notes, I listed a collection of statistics that every neuroscientists should know in order to perform back-of-the-envelope calculations. The Harvard BioNumbers site provides a searchable database of useful numbers. For instance, I recently wanted to know about nucleotide misincorporation rates and this query provided me with the information I was looking in writing for this post. There are similar on-line resources available for many scientific and engineering disciplines offered by academic institutions and professional societies, and it is well worth your effort to ask an expert for a credible referral and track them down.
Cellular transcoding processes such as DNA replication and messenger RNA transcription make occasional errors incorporating nucleotides into their resulting products. Zamft et al [173] propose a method of exploiting this property to build a molecular recording device. The authors note that the rate of misincorporation is dependent on the concentration of cations, specifically Ca+2 ions. They show that by modulating cation concentrations one can influence the misincorporation rate on a reference template in a reliable manner so that information can be encoded in the product of DNA polymerase and subsequently recovered by sequencing and comparison to the reference template.
The paper also includes a statistical analysis of misincorporation events, a Bayesian method for calculating the Shannon information gain per base pair, and an estimate of recording media storage capacity — depending on the particular polymerase and cation modulation rate, they estimate 11 megabytes could be stored on a template the length of a human genome (3.2 × 109 bases). In principle, this approach could achieve high temporal resolution combined with very high spatial resolution. This HHMI animation of DNA replication provides a dramatic illustration of the potential power of harnessing DNA polymerase in this fashion. Adam Marblestone — a graduate student in George Church’s lab and one of the paper’s co-authors — has analyzed the efficiency of their approach in terms of its energy requirements and processing throughput compared with a CMOS solution. Adam will be talking about his analysis in class later in the quarter.
March 29, 2013
A new BAM paper [5] was just published (PDF) focusing on potential recording and readout solutions based on advances in nanotechnology. We’ll be covering this paper in class and so I’ll put off reviewing it here. But in reading the paper I ran across this idea: “non-optical methods that leave a recoverable trace of activity within cells to side-step the light-scattering problem, which could involve gene encoding a designer polymerase transduced into a genetically targeted subset of neurons, especially if the polymerase were engineered for increased error rate in elevated Ca2+ which can track neural activity patterns at high speeds even in the nucleus”, which included a reference to a 2011 paper by Konrad Kording [88] with the intriguing title “Of Toasters and Molecular Ticker Tapes”. Not more than a couple of hours after I read this, Kording sent me email out of the blue saying that he’d seen the transcript of the Berkeley talk and suggested I might want to take a look at another paper of his [142].
In this paper, Stevenson and Kording discuss some of the implications of advances in neural recording with respect to data analysis (PDF). Having surveyed some of the limitations with current solutions to the readout problem, the authors opine:
Despite these limitations, whole-brain spike recordings may not be beyond the realm of possibility. For example, one might imagine a system in which each neuron records spike times onto RNA molecules that could then by read-out by sequencing the results, one neuron at a time. Just as microchip fabrication technology has evolved drastically since the introduction of Moore’s law, progress in neural recording technology may allow growth beyond our current expectations.In the earlier work [88] referenced in the BAM paper, Koerding refers to this idea as a molecular ticker-tape after first mentioning Tony Zador’s work [172] on sequencing the connectome by encoding connectivity patterns in DNA sequence barcodes. Once we have the connectome, Kording suggests the next step is to record neural activity:
Lastly, it seems that the step of recording neural activity can also be reduced to DNA sequencing. When a cell divides, it naturally copies its entire DNA using DNA polymerase. The movement of the polymerase along the DNA template could be engineered to be essentially a molecular ticker tape, such that the environment at that point in time is recorded in the DNA sequence. This could be achieved by engineering a polymerase that would make errors when neural activities are high, for example, such errors could be modulated by calcium concentration. While copying a template, DNA polymerase could thus write the temporal trace of activity as error patterns onto DNA molecules. Of course, these would be difficult steps, and neither DNA polymerase that depends on neural activity nor steady template copying in quiescent neurons has been established. Still, the sketched approach could in principle allow high temporal resolution combined with very high spatial resolution.To record information in DNA, Kording refers to a patent by George Church and Jay Shendure for a “Nucleic Acid Memory Device” which appears worth a closer look from my cursory reading.
Here is an update on recent developments in synthetic biology, Drew Endy’s latest exploits, and the BIOFAB facility which he co-directs. In a related story, a paper in Science Endy and his colleagues at Stanford describe a synthetic-biology device they call a transcriptor which provide an analog transistor for building biological lagic gates:
Organisms must process information encoded via developmental and environmental signals to survive and reproduce. Researchers have also engineered synthetic genetic logic to realize simpler, independent control of biological processes. We developed a three-terminal device architecture, termed the transcriptor, that uses bacteriophage serine integrases to control the flow of RNA polymerase along DNA. Integrase-mediated inversion or deletion of DNA encoding transcription terminators or a promoter modulate transcription rates. We realize permanent amplifying AND, NAND, OR, XOR, NOR, and XNOR gates actuated across common control signal ranges and sequential logic supporting autonomous cell-cell communication of DNA encoding distinct logic gate states. The single-layer digital logic architecture developed here enables engineering of amplifying logic gates to control transcription rates within and across diverse organisms. (source)
There’ve also been some interesting developments in terahertz imaging [124] reported in this news article. Unfortunately, the prospects for increased penetration depth in biological samples are not appreciably brighter in the near term. This survey article on terahertz imaging provides a sobering outlook for the roll out of this and related technologies: The authors do address the question of tissue penetration in biological applications; however, they believe that despite their drawbacks in terms of, respectively, radiation and relatively low resolution, the incumbent X-ray and MRI technologies will provide stiff competition and therefore companies will target improving terahertz technology in areas where this technology really shines, e.g., reduced scattering leading to improved resolution.
March 27, 2013
David Heckerman sent around this video showing off the MRI-guided ultrasound technology being developed by Insightec. This is the company Dr. Kassell at FUSF and the engineers at Siemens were telling us is currently conducting patient trials for their essential-tremor therapy. I’m assuming that the MRI is used to guide the focused-ultrasound-ablation scalpel and adjust the beam to compensate for the variable thickness of the skull. I wouldn’t have thought that possible, but perhaps they’re using some new NMR techniques and contrast agents to create a good-enough model of the cranium using the spatial resolution of the MRI. Or it could be that the size and positional accuracy of the beam is not as critical in their applications, and so they don’t have to resort to CT to build an accurate 3-D model or they use feedback to control the array of transducers. In any case, at least the MRI doesn’t have the radiation problems associated with CT.
CORRECTION: It turns out that the InSightec system — called ExAblate — does rely on a pre-operative CT scan that likely would incur some amount of radiation exposure. An InSightec white paper describes the system and current protocols for functional neurosurgery and tumor ablation, stroke, and targeted drug delivery. The CT scan is only required in the neurosurgical protocol to construct an initial model of the skull which is subsequently registered using MRI once the patient is placed in the ExAblate with his or her head immobilized in a stereotactic frame28. I tracked down several related methods [67, 102] some of which start with a CT scan and use MRI for subsequent registration and others that avoid the CT scan altogether29. Understanding the implications of this technology for readout applications will take some time and having the patient immobilized in an MRI scanner is obviously not the optimal context for studying the relationship between behavior and brain activity, but if this lives up to the claims, it does appear to offer a promising component technology for addressing the readout problem.
March 25, 2013
For my technical talk at Google on Monday, I decided to try a new tack for introducing and motivating the problems facing the Brain Activity Mapping Project. Here’s the new introduction:
Among the many different types of brain activity one might want to record, action potentials or spikes are high on most neuroscientists wish list. An action potential corresponds to an abrupt change in the electrical potential across the cell membrane initiated in the axon hillock30 near the neuron cell body or soma and propagated along the axon to the synaptic terminals. Researchers would also like gene expression levels, ion concentrations, neurotransmitter indicators, and a host of other observations, but we’ll focus on spikes for this discussion.You can find a full-length version of the action-potential animation I used in the talk here and an unedited version of the animation illustrating synaptic transmission at a chemical synapse31 here (Source: W.H. Freeman). Someone asked me if there were synapses on axons and I replied that the rule in biology is that if it can be done then nature has probably done it, and, in this case, the answer is definitely yes. Axons terminating on the postsynaptic cell body are axosomatic synapses. Axons that terminate on axons are axoaxonic synapses. I also used some clips from the “Inner Life Of A Cell” video produced by BioVisions at Harvard. If you want to see just the clips I used, fast forward 3:20 into the video for microtubules, transport vesicles, kinesins (motor proteins), and 4:40 for the clip on ribosomes, ribonucleic acids, ribonucleotides and amino acids.Once the membrane potential reaches the threshold for spike initiation, a spike is likely to soon follow and propagate along the axon to the synapses. If we could locate a nanoscale sensor in the axon hillock to record the local membrane potential that could be quite useful. In fact, it would probably be almost as useful to simply record when the potential exceeds the threshold.
There are other types of activity correlated with spikes that may turn out to be easier to record. When a signal is transmitted across a synapse from one neuron to another, calcium ions play a key role in the release of small packets or vesicles containing neurotransmitters. By recording the concentration of calcium ions in the synapse, we obtain a lagging indicator for synaptic transmission and thus the propagation of information from cell to another.
We have the technology for recording membrane potentials and calcium concentrations, but reading off these signals at scale is potentially problematic. The sensors are essentially biomolecules that fluoresce when certain local states obtain. The patterns of fluorescence are read off by a very sensitive optical imaging device called a two-photon excitation microscope.
However, a two-photon microscope relies on light and, while it is able to penetrate deeper than conventional light microscopes — see Helmchen and Denk [66], the depth of penetration for state-of-the-art technology is limited to at most a few millimeters. BAM researchers would like to read off such information for millions of neurons simultaneously at millisecond resolution in an awake behaving human subject.
March 23, 2013
Greg Corrado, Jon Shlens and I met with David Cox at Google on Wednesday to catch up on what’s going on David’s lab and, in particular, to learn about the robotic microelectrode placement work that he’ll be talking about when he visits class on May 22. We talked about the Brain Activity Mapping initiative and I asked him what would be on his wish list for scalable neural-state readout. He was most interested in spike timing data and its correlates. We talked about improvements in calcium imaging GCaMP technology and what’s possible now that GCaMP6 is available. In looking for GCaMP-related references for my class, I came up with this paper32 from Janelia by Akerboom et al [73] discussing their experiments with the current crop of genetically encoded calcium indicators including a family of GCaMP5 fluorescent sensors.
We also discussed innovation on the imaging side as he is soon to take delivery of a new two-photon microscope which he intends to modify for his purposes. The literature is full of researchers hacking their microscopes, but a paper by Grewe et al [60] caught my eye both for their claims about temporal and spatial resolution and the discussion of methods for accelerated scanning. They modified their microscope with an axial scanning device that employs chirped acoustic waves traveling through acousto-optical deflectors that “act as an acousto-optical lens to control beam divergence in addition to deflection angle, resulting in a movement of the excitation spot along the z-axis.” They claim33 to have achieved “near-millisecond” precision using this method.
After a discussion of the tradeoffs involved in calcium imaging, we talked about recording action potentials and the state-of-the-art in voltage-sensitive dyes. As possible target goal for new BAM recording technology, we compromised on the following — still quite ambitious — requirement for a scalable spike reader: for each neuron, record the voltage at its axon hillock at millisecond resolution, with a simplifying binary “spike” versus “no spike” signal as a further compromise. Regarding state-of-the-art in voltage-sensitive dyes, I found lots of pointers to Dejan Zecevic’s lab at Yale including this Popovic et al [99] paper34. David mentioned recent news on voltage-sensitive dyes also from Yale and suggested we look into the work of Vincent Pieribone — who by the way sounds like an interesting fellow. This Barnett et al [12] paper35 seems the closest match to what David was telling us about. I haven’t read them but these two additional papers from Pieribone’s publication page also looked interesting:
Jin, L. and Han, Z. and Platisa, J. and Wooltorton, J. and Cohen, L. and Pieribone, V.A. (2012) Single action potentials and subthreshold electrical events visualized in neurons using a fluorescent protein voltage sensor. Neuron 75:779-85.
Baker, B. J. and Jin, L. and Han, Z. and Cohen, L.B. and Popovic, M. and Platisa, J. and Pieribone, V. A. (2012) Genetically encoded fluorescent voltage sensors using the voltage-sensing domain of Nematostella and Danio phosphatases exhibit fast kinetics. Journal of Neuroscience Methods 208:190-6.
P.S. I asked David if the Grewe et al [60] paper did a good job summarizing the state-of-the-art for spatial and temporal resolution in recovering spike timing from calcium imaging and he answered as follows:
[T]his paper is a reasonably fair representation. At some point, if the spike rates are high enough, the temporal characteristics of the calcium transient start cramping your style, but you can do some reasonable reconstruction of lower-rate spike trains as long as you’re willing to give up some FoV or give up continuous imaging. Emerging fast voltage sensitive fluorophores should enable this kind of game to be played even more effectively (and might give us access to subthreshold stuff), assuming the SNR’s keep increasing. There’s an awful lot of structure in spike trains and brain images, so I think there’s a lot of room to exploit that structure to read-out brain signals more efficiently.
March 21, 2013
The physics of fluorescence and the use of fluorophores play important roles in neuroscience and biology more generally. If you’re unfamiliar with these concepts, this short tutorial should serve to get you started. The following Jablonski diagram describes the basic process whereby a fluorophore absorbs a photon causing an electron in the outer shell of its valence band to jump the band gap and temporarily occupy a shell in the conduction band before falling back to its original energy state and emitting a photon:
 |
As a first approximation, think of quantum dots as fluorophores corresponding to inorganic molecules. Their main advantage over organic fluorophores is that they can be manufactured to precise specifications and so we aren’t limited to what we can find in nature. Their main disadvantage is that the materials from which they are made are not soluble in water and in many cases are toxic to cells. For an introduction to the application of quantum dots accessible to students with high-school physics and biology see Walling et al [161]. The following graphic from [161] illustrates the primary characteristics of quantum dots used for in vivo imaging:
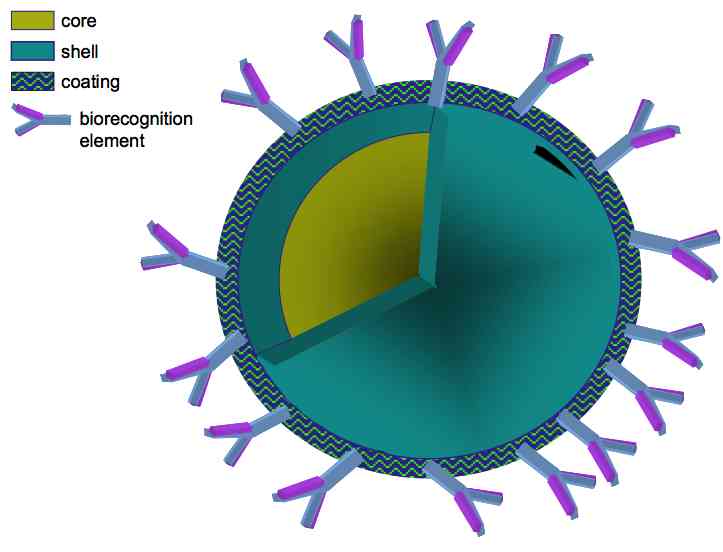 |
Fluorophores are subject to destruction by photobleaching which engineered fluorophores like quantum dots can be designed resist. The Walling et al [161] paper also has a good introduction to flow cytometry using quantum dots as a method for cell counting, sorting and biomarker detection — see Pages 470, 471 and 477. In discussing the problem of quantum-dot toxicity, the authors mention silicon bicarbide36, which is chemically inert, stable and biocompatible, as a promising alternative — see Page 473.
Fluorescence resonance energy transfer or FRET is a mechanism — analogous to electromagnetic near-field communication — describing the energy transfer between two chromophores: the donor and acceptor. When the donor absorbs a photon it either fluoresces emitting a photon or it emits a virtual photon that is absorbed by the acceptor thereby transfering energy. The probability that a photon absorbed by the donor will result in an energy transfer is a function of the spectral overlap of the donor emission spectrum and the acceptor absorption spectrum and the distance separating the two fluorophores. If the acceptor absorbs enough energy it will fluororesce. Using a measure called the FRET efficiency it is possible to apply FRET to measure very small distances. Here is the Jablonski diagram for at FRET pair:
 |
March 19, 2013
Jon Shlens pointed me to the work of Alan Jasanoff, a biophysicist and faculty member in MIT’s Department of Biological Engineering. Jasanoff is developing a new class of contrast agents for nuclear magnetic resonance imaging that he hopes will considerably improve spatial and temporal resolution and record neural correlates such as calcium concentrations that are more highly correlated with brain function. This article about him has a nice characterization of the challenges he faces:
Designing a contrast agent is difficult — a challenge comparable in many ways to designing a new drug. First, the agent must bind reversibly to a target molecule that is directly related to brain activity. It must also contain a so-called paramagnetic metal (such as iron or manganese) that changes the agent’s magnetic properties when it binds to the target molecule, so that it produces a different MRI signal depending on whether the target molecule is present or absent. The agent must also be stable within the brain and must not interfere with normal brain function. And if the contrast agent is used to detect changes that occur inside neurons, it must cross the surface membrane that separates the neuron from its surrounding environment.The article goes on to describe Jasanoff’s work on imaging dopamine as well as metals such as calcium and zinc that are correlates of brain activity. The research is still in the early stages but there are some publications on his website showing promising preliminary results.
Yael Maguire suggested a paper by Zamft et al [173] as a promising first step toward a chip for deep-sequencing39 within complex tissues such as would be required in functional neural connectomics. George Church and Ed Boyden are co-authors on the paper.
I was telling Mark Segal about the Brain Activity Mapping Project and he was reminded of a scene from the The President’s Analyst in which James Coburn talks with someone from the phone company who wants the congress to pass a bill requiring all babies to have a miniature phone called a “Cerebrum Communicator” implanted in their “anterior central gyrus” which the phone company representative tells the Coburn character “is simply that portion of the brain in which intellectual associations take place.” Billing would take place automatically through a communication tax paid directly to the phone company. I thought this might make a nice addition to my Berkeley talk. You can check it out in this YouTube video.
March 17, 2013
I’ve seen several articles lately touting the advantages of quantum-dots (QD) over traditional organic fluorophores, e.g., Zhang, Y. and Wang, T.H. Quantum Dot Enabled Molecular Sensing and Diagnostics. Theranostics 2(7):631-654 (2012). This review by Daniel Levine that I found on a random MIT website provided a surprisingly good start for my investigations. I was particularly interested in recent papers describing QD-FRET technologies for sensing membrane potentials40, despite repeated statements of concern about QD toxicity.
This led me to do a literature search on QD toxicity and read a couple of reviews. Cadmium-based QDs seem unlikely to serve for anything other than short-duration in vivo experiments in which the target cells can be sacrificed. Indium phosphide QDs appear to be less toxic but it is too early to tell whether they will replace cadmium-based QDs in animal studies any time soon. Nonetheless these new QDs are intriguing and probably bear further investigation. Here are the relevant references:
Walling et al [2009] provide a good review of current QDs for live-cell imaging including details on QD encapsulation using ZnSe to mediate some of the toxic effects of cadmium-based QDs. Source: Quantum Dots for Live Cell and In Vivo Imaging. Maureen A. Walling and Jennifer A. Novak and Jason R. E. Shepard. International Journal Molecular Science. 10(2):441-491. (2009).
Valizadeh et al [2012] summarize the results from a dozen or studies on toxicity and relate work suggesting that “QDs could not only impair mitochondria but also exert endothelial toxicity through activation of mitochondrial death pathway and induction of endothelial apoptosis” and the “cytotoxicity of CdTe QDs not only comes from the release of Cd2+ ions but also intracellular distribution of QDs in cells and the associated nanoscale effects.” Source: Quantum dots: synthesis, bioapplications, and toxicity. Alireza Valizadeh and Haleh Mikaeili and Mohammad Samiei and Samad M. Farkhani and Nosratalah Zarghami and Mohammad Kouhi and Abolfazl Akbarzadeh and Soodabeh Davaran. Nanoscale Research Letters 7(1):480-494 (2012).
Chibli et al [2011] consider the toxicity of InP/ZnS QDs which appear to have significantly lower toxicity than Cd based QDs but are still far from being biofriendly, noting “Indium phosphide quantum dots have emerged as a less hazardous alternative to cadmium-based particles [...] Although their constituent elements are of very low toxicity to cells in culture, they nonetheless exhibit phototoxicity related to generation of reactive oxygen species by excited electrons and/or holes interacting with water and molecular oxygen.” Source: Cytotoxicity of InP/ZnS quantum dots related to reactive oxygen species generation. Hicham Chibli and Lina Carlini and Soonhyang Park and Nada M. Dimitrijevic and Jay L. Nadeau. Nanoscale 3(6):2552-2559 (2011).
The 2013 paper by Brunetti et al is more encouraging but it may be some time before we have the data necessary to make any predictions about their possible use in human subjects. Source: InP/ZnS as a safer alternative to CdSe/ZnS core/shell quantum dots: in vitro and in vivo toxicity assessment. Brunetti, V. and Chibli, H. and Fiammengo, R. and Galeone, A. and Malvindi, M.A. and Vecchio, G. and Cingolani, R. and Nadeau, J.L. and Pompa, P.P. Nanoscale. 5(1):307-317 (2013).
Follow-up discussion included this question from Ed Boyden:
ESB: Safety is an analog variable, of course. Over sufficiently long timescales, and at high enough doses, almost anything can be toxic. I was under the impression that QD coatings were getting to be quite good, no?If you’re doing any bio-related literature searches, you might want to check out the BioInfoBank Library as it makes it easy to search, view abstracts and follow citation paths backward and forward. The BibTeX entries are reliably formatted and consistent so that you can update your bibliographic database and add a citation to a mail message or log entry with a single command. It is better as a first stop than CiteSeer or National Center for Biotechnology Information (NCBI) for papers relating to biotechnology, though I generally consult the National Library of Medicine PubMed resources as well.TLD: There has been a lot of work developing surface coatings to address such issues as solubility, stability and conjugation. For example Walling et al [161] describe using a surface coating consisting of dihydrolipoic acid (DHLA) for coordination to the quantum dot surface, a short polyethylene glycol (PEG) spacer for increased solubility and stability, and reactive carboxyl or amino functional groups for conjugation.
Surface coatings also serve to reduce toxicity. The release of cadmium ions from the CD core has been implicated as the primary cause of toxicity41, and so surface coatings such as PEG that reduce or eliminate oxidation are key to reducing toxicity by isolating the core. ZnS is another common surface shell coating for CdSe core QDs which has been shown to aid in reducing cytotoxicity.
In addition to cell death caused by cadmium ion release, QDs can also damage DNA and disrupt normal cell activity caused by factors such as the surface coatings themselves. In one set of experiments involving human fibroblast cells, PEG-silica-coated QDs were shown to impact the expression levels of only about 0.2% of all the genes studied. However, DNA damage was observed in experiments using QDs coated with carboxylic acids.
Walling et al [161] conclude by stating: “Because of the limited information on quantum dot cytotoxicity and the continued ongoing research, broad generalizations regarding quantum dot cytotoxicity should be forestalled, until conflicting reports on surface coating protection and degradation, core oxidation, cell activity disruption, and toxicity are reconciled and additional conclusive studies have been performed and validated.”
March 13, 2013
Here’s a simple question crying out for a back-of-the-envelope calculation: How fast are we spinning around the earth’s axis? And here’s my back-of-the-envelope calculation: It’s about 2.4K miles across the US. I’d say you could string about 10 of the US end-to-end around the globe yielding an estimate of 24K in circumference. Divide by 24 hours and you get 1,000 MPH. Let’s check on the off-the-top-of-my-head approximations that I used: With a little help from Google we find out that the driving distance between New York and San Francisco is approximately 2,906 miles, the straight-line distance from New York to San Francisco is about 2582 miles (4,156 km). In addition, the diameter of the Earth is around 7,918 miles (12,742 km) and its circumference is approximately 24,901 miles (40,075 km).
A more precise phrasing of the original question would be the following: What is the linear velocity of a point resting on the equator assuming that the earth is perfectly spherical? And here is a high-school-physics student’s calculation: Starting with a little help from Yahoo Answers we get the time period of the Earth T = 23 hours, 56 minutes, 4 seconds long or 86,164 seconds, and, given that it rotates through 2 π radians in this time, the angular velocity is ω = 2 π / T = 7.2921 * 105. The radius of the earth is R = 6.38 * 106 meters and so the linear velocity V = R * ω = 465.25 meters per second. Checking the math and rounding the result, we get 6.38 * 106 * 2 * π / 86164 = 465 as a sanity check, and converting the result to MPH we have 602 * 465.25 / 1609.34 = 1040.79 MPH. Our back-of-the-envelope calculations wasn’t off by much in this case, but it could be off by plus or minus 10% and it probably still would have been fine for our purposes42. Now see if you can appreciate Chris Uhlik’s quick-and-dirty, back-of-the-envelope analysis of the prospects for solving the readout problem using more-or-less conventional RF technology.
March 11, 2013
There was a Stanford news release this morning featuring Karl Deisseroth and pointing to a short article in Science: The Brain Activity Map, A. Paul Alivisatos and Miyoung Chun and George M. Church and Karl Deisseroth and John P. Donoghue and Ralph J. Greenspan and Paul L. McEuen and Michael L. Roukes and Terrence J. Sejnowski and Paul S. Weiss and Rafael Yuste, Science, March (2013). The article includes several authors of the 2012 Neuron paper [6] and serves primarily as a challenge to the ACS community. It did include one new reference for me that has a tempting title: Nano in the brain: nano-neuroscience. A. M. Andrews and P. S. Weiss. ACS Nano 6:10 8463 (2012). Unfortunately, the reference is mostly gratuitous with a tease of a title and very little substance to warrant it. I scanned the abstracts and more than 90% have nothing to do with BAM challenges. Which is not say there aren’t a few articles of possible interest, but here the only ones that I could see even scanning:
The State of Nanoparticle-Based Nanoscience and Biotechnology: Progress, Promises, and Challenges
Colloidal Quantum Dots as Saturable Fluorophores
Smartly Aligning Nanowires by a Stretching Strategy and Their Application As Encoded Sensors
DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties
Fluorescent Polymer Nanoparticle for Selective Sensing of Intracellular Hydrogen Peroxide
March 9, 2013
There was a story in The Economist mentioning the Brain Activity Mapping Project, but focusing primarily on the researchers working on the Human Connectome Project (HCP), specifically Jeff Lichtman, of Harvard University and Steven Petersen at Washington University, in St Louis. Lichtman employs serial-section microscopy to tissue samples as in the work of Briggman [24], Denk [8], and Seung [74]. Petersen uses functional connectivity MRI (fcMRI) to track water molecules — this method is related to diffusion tensor imaging techniques that we encountered in tracing myelinated axon bundles in cortex — in awake human subjects to map out correlations between the metabolic activity of different brain areas. If you’re unfamiliar with their work, you might want to check out the (above) links to their labs. Also see here for summary description of new a HCP dataset.
A second article takes a broader view of the grand-challenge goals of the BAM Project and suggests that it may be too ambitious. It also mentions in passing that all of the five authors of the paper in Neuron are well positioned to apply for funding if the program moves forward and the lead author Paul Alivisatos, director of the Lawrence Berkeley National Laboratory, is eager to take on the nanoscale instrument builder role. Motivations aside a critical assessment is warranted and no one has seriously taken on this challenge. The student projects in CS379C could help in this assessment.
According to a recent article, a class of human glial cells called astrocytes when transplanted into mice apparently influences communication within the brain, allowing the mice to learn more rapidly. According to the article, “[a]strocytes are far more abundant, larger, and diverse in the human brain compared to other species. In humans, individual astrocytes project [to] scores of fibers that can simultaneously connect with large numbers of neurons, and in particular their synapses, the points of communication where two adjoining neurons meet. As a result, individual human astrocytes can potentially coordinate the activity of thousands of synapses, far more than in mice.” If the results are replicated, this could radically change our theories on neural function and neurodegenerative diseases such as Alzheimer’s. Another puzzling study by researchers at Oxford reveals that transcranial electrical stimulation can “enhance learning when they targeted a certain spot. But that also made people worse at automaticity, or the ability to perform a task without really thinking about it. Stimulating another part of the brain had the reverse effect, on both learning and automaticity.” Along similar lines, Yale researchers found a gene normally turned off in adult mice which when turned back on had the effect of reseting the older brain to adolescent levels of plasticity.
March 7, 2013
Here is another example of using eye tracking to diagnose head injury. This article in USNews summarizes a research paper in the April issue of the journal Stroke describing a diagnostic device — known as a video-oculography machine – which is a modification of a “head impulse test” used regularly for people with chronic dizziness and other inner ear-balance disorders. “This is the first study demonstrating that we can accurately discriminate strokes and non-strokes using [a pair of goggles that measures eye movement at the bedside in as little as one minute]”, said Dr. David Newman-Toker, lead author of the research paper.
March 5, 2013
Here are some videos, talks and web pages that I watched, listened to or read over the last couple of days but have no time to summarize and so I list them here so they won’t get lost: Ed Callaway’s McGovern Symposium video on rabies-based tools for elucidating neural circuits and linking connectivity to function. The MIT McGovern Institute Symposium has a bunch of short educational videos on brain science including one video on the MIT and Georgia Tech automated-patch- clamping robot and a second showing an animation of optogenetics — each video runs less in than five minutes. Brainbow 3-D neural-circuit reconstruction featured in a YouTube video. MIT Professor Robert Langer on Science Friday podcast includes a discussion of how his remarkable lab has remained so successful over several decades. The list of projects on his lab research page lists a number of critical enabling technologies for BAM. This Google website describing how search works provides a quick introduction to how scaling works at Google. A discussion with George Church on the brain activity mapping project. Here is a link to Mark Schnitzer’s startup where you can learn more about their hardware for streaming live HD video of large-scale neural activity in naturally-behaving mice. Violin Memory’s business solutions make for an interesting look at the company: “Removing barriers to big data with flash memory arrays”, “How flash memory arrays can help enterprises expand the scale of their datasets”, and “Process online transactions 20x faster with less latency and achieve the lowest cost per transaction”.
March 3, 2013
Here is our strategy for making CS379C a more valuable experience for both the students and our outside participants and invited speakers: We want to get the best students to participate in CS379C. We want to motivate them to come up with the best ideas and generate the most interesting proposals. Some of the students are interested in pursuing careers in science and others want to found a successful startup. These two goals don’t have to be at odds with one another. We’re putting together a stellar group of scientists, engineers and entrepreneurs to suggest topics and provide students with advice on the relevant areas of biology, neuroscience, nanotechnology and industrial-scale computation and information retrieval. If we are the only audience, then the impact of their work will have primarily localized impact, unless they take the initiative to promote their ideas more widely. Why shouldn’t we provide that extended audience and make their project presentations include a pitch to a blue-ribbon panel of scientists, foundation directors and venture capitalists.
We know a lot of players in this space: scientists, engineers, foundation directors, investment firm partners and program managers from NIH and NSF, Salk Institute, Cold Spring Harbor Laboratory, Siemens, Focused Ultrasound Foundation, Janelia Farm Research Campus of HHMI, Google Ventures, plus a host of colleagues from great research universities. Those we can’t tempt to attend “The Great Brain Readout” in person or by VC at Google, might be interested in reviewing a nicely edited video featuring the best pitches as voted by those attending “The Pitch” event. Internally, we can probably get Alan Eustace (Google SVP Engineering), Alfred Spector (Google VP Knowledge), Astro Teller (Captain of Moonshots Google X), Ray Kurzweil (Google Director of Engineering), Bill Maris (Google Ventures), and Krishna Yeshwant (Google Ventures) interested enough to at least look at the video of top-rated pitches. If BAM is funded, I’ll bet NSF and NIH would be interested in take a close look at the best projects. There might even be a publication or funding in it for the students with the best ideas.
March 2, 2013
Here is a set of topics for CS379C projects along with one or two possible consultants for each topic to help with identifying relevant papers and offering advice. These suggestions are primarily concerned with possible solutions to the “readout” problem — just conveying information out of the brain, however, we will consider topics relating to the “sensing” problem — how to record neural-state information including membrane potential, protein expression, calcium concentration and their correlates, and the “inference” problem — suppose we were able to record spikes or protein expression levels from millions of locations within a neural tissue at millisecond resolution, how might we make sense of this deluge of data43:
Assessment of tissue damage caused by ultrasound scanning plus basic calculations for back-of-the-envelope analyses — Neal Kassell, Arik Hananel
Photoacoustic imaging with NIRS44 illumination delivered by a relatively sparse invasive network of fibre optic cables — Ed Boyden, Carl Deisseroth
Automatic registration of tissue samples with a set of standard exemplars spanning the space of natural variations — Mike Hawrylycz, Clay Reid
Whole brain mapping of Brodmann areas or alternative anatomic and cytoarchitectural landmarks and areas of interest — Larry Swanson, Fritz Sommer
Proposal for scaling array tomography to cover full gamut of synaptic proteins to complement Allen Mouse Brain Atlas — Stephen Smith, Mark Schnitzer
Analysis of the sequencing approach with focused ultrasound raster scan with single and multiple beams and reporters — Bruno Madore, Ben Schwartz
Prospects for building brain-wide self-organizing nanoscale communication networks for solving the readout problem — Yael McGuire, Akram Sadek
Analysis of the bandwidth and frequency-spectrum issues involved in a cellular-radio-model for whole brain readout — Chris Uhlik
Prospects for growing neural networks or brains around a scaffolding implementing fiber optic communication network — Ed Boyden, Greg Corrado
Computational strategy for managing the billions of terabytes of SEM45 data likely generated by scalable BAM technology — Mike Hawrylycz, Luc Vincent
Proposal for scaling SEM based methods to produce detailed connectomic maps to complement Allen Mouse Brain Atlas — Kevin Briggman, Sebastian Seung
Analysis of proposal in the “Sequencing the Connectome” paper [172] with completion date based on off-the-shelf technology — Tony Zador, Jon Shlens
March 1, 2013
This morning I was on the phone with Dr. Arik Hananel and his colleagues at the Focused Ultrasound Surgery Foundation (FUSF) where Arik is Scientific and Medical Director. I wanted to know more about using FUS to treat essential tremor — see here and here for news releases. In addition to Arik, we were joined by Dr. Neal Kassell who is the chair of the foundation, Jessica Foley, one of the foundation’s resident scientists, and John, the technical lead for the brain program. We talked a little about FUS-beam-focus size and collateral tissue heating and cell damage, but spent most of the time discussing the foundation, its mission and the prospects for new treatment methods and additional clinical trials. The FUSF website has a lot of useful resources, and Arik and Neal offered to help out in digging up relevant papers and making connections with their industry and academic partners. I found their publications page a good place for browsing the latest FUS technology papers. Neal told me about their (relatively) new brain program emphasizing MRI-guided FUS. The white paper that resulted from their 2011 Brain Workshop mentions a lot of issues I’d like to know more about, and I’ve asked Dr. Kassell for some relevant review articles.
Sometimes it seems that doctors are even more aware of status and credentials than most academics and have a dismissive attitude to those outside the medical profession dabbling in their business, but Arik and Neal were generous with their time and seemed genuinely interested in BAM and our related interests. Neal perked up when, during a discussion of entrepreneurial physicians, I told them that my Uncle Frank is Dr. Frank Mayfield, the neurosurgeon who invented the spring aneurysm clip and the Mayfield skull clamp and founded the Mayfield Clinic in Cincinnati. These two devices are constant companions to practicing neurosurgeons and, while they may be as common as paper clips and stocking caps in my world, both Arik and Neal were aware of and grateful to the person who invented them. Jessica didn’t say much, but I noticed several of her papers when I did a web search, including this one: Effects of high-intensity focused ultrasound on nerve conduction. Jessica L. Foley and James W. Little and Shahram Vaezy. Muscle & Nerve, Volume 37, Issue 2, pages 241-250 (2008). (URL). Here’s the note that I sent to Arik and Neal with a follow-up question:
Thanks for the discussion this morning. I spent a couple of hours looking through the FUSF web pages — lots of great resources. In particular, the library of relevant papers is very useful and I’ll definitely link to it for my Stanford class in the Spring. I sent email to a couple of partners in Google Ventures to see if I can interest them in the general FUS area and FUSF-related interests in particular. Hopefully I’ll get a chance this weekend to go through my meeting notes and process what I learned today. I did have one question / request that you might be able to help me with: The list of issues raised in the white paper for the 2011 Brain Workshop were tantalizing, but left me with more questions than I started with. Could you recommend a paper reviewing what is known about potential cell-damage associated with diagnostic uses of ultrasound and FUS-based surgical interventions outside of the beam focus?The Siemens engineers mentioned some rough back-of-the-envelope numbers relating to temperature monitoring using MRI, intensity measurements for diagnostic US, and cumulative consequences measured in terms of spatial peak-temporal average intensity and mechanical index, but I’d like to understand both the physiological issues and the methods for calculating such measures. In particular, I’m interested not just in the power delivered to the focus but the cumulative effects of energy absorbed and scattered outside the focal area. The feasibility of the scanning approach I sketched will depend on the ability to maintain the tissue temperature within a narrow band around some normal time-varying operating temperature throughout repeated scans. The nano-reporters will have to be sensitive to the temperature / pressure derivative rather than its absolute value since the latter will vary with behavior. I should reiterate that this approach is only one of many we’re looking at.
February 27, 2013
Always on the lookout for alternative uses of existing technologies, I ran across this paper46 by Wang et al [163] discussing focused ultrasound using microbubbles to modify the permeability of the blood-brain barrier and photoacoustic microscopy using gold nanorods47 that are selectively excited by a tunable laser to monitor permeability. The primary objective is to monitor focused-ultrasound-induced blood-brain barrier opening in a rat model in vivo. The idea that focused-ultrasound (FUS) can induce changes in the blood-brain barrier (BBB) that would facilitate the passage of nanoparticles to deliver drugs is interesting in and of itself. One problem with this method is that the degree of BBB opening varies substantially and it is important to be able to monitor changes in the BBB in order to control dosage. Currently such monitoring is done with contrast-enhanced MRI, but the spatial and temporal resolution of this method is a limiting factor.
The procedure described in the paper is a little complicated but I’ll try to provide a succinct summary with additional detail in footnotes excerpted from the paper. First, the FUS method of induced BBB opening is applied — the rat undergoes a craniotomy to aid in beam focusing and power modulation, and then, prior to sonication, an ultrasound contrast agent (SonoVu [49] developed by Bracco Diagnostics) is intravenously injected to facilitate acoustic cavitation. The details regarding power and pulse length provide interesting insight into how to control for adverse effects in the test animals. Next AuNRs48 are injected into the jugular vein and accumulate at the BBB opening foci following sonication. Finally, dark-field confocal photoacoustic microscopy is applied for monitoring the BBB opening. The area to be monitored is illuminated by a tunable laser49 and then scanned by an ultrasonic transducer using a set of piezoelectric drive motors with a step size of 120μm in each of two directions.
In the conclusion section of the paper, the authors claim that the “experimental results show that AuNR contrast-enhanced photoacoustic microscopy successfully reveals the spatial distribution and temporal responses of BBB disruption area in the rat brains. The quantitative measurement of contrast enhancement has potential to estimate the local concentration of AuNRs and even the dosage of therapeutic molecules when AuNRs are further used as nanocarriers for drug delivery or photothermal therapy.”
Reading the Wang et al [163] paper and, in particular, the part about photoacoustic imaging using infrared laser and gold nanorods, got me interested in the limitations of near-infrared spectroscopy (NIRS). NIRS devices are used as sensors in gaming headsets and increasingly as alternatives to MRI and CAT for infants. NIRS variants have been applied to both detect breast tumors and guide surgeons in performing biopsies allowing imaging at depths exceeding 3cm [83]. Work with pulsed lasers and quantum-dot contrast agents continues to push the envelope but I was wondering to what extent the penetration and power dissipation issues can be overcome except perhaps by using a relatively sparse — but invasive nonetheless — fiber optic network to deliver light to deeper tissues. This short introduction to the issues involved in light penetration in living tissue provides some insight.
Benjamin Schwartz from Bruno Madore’s lab now at Google told me about Kim Butts Pauly’s lab at Stanford and her work on applying FUS to the treatment of metastatic bone tumors for the palliation of pain50.
February 25, 2013
Here’s Bruno Madore’s response to my question about using his technique [100] for focused scanning with multiple beams:
BM: Your email is intriguing, especially the part about a “nanoscale reporter device”. For sure, if you can create sparsity through the use of a limited number of such devices, there may very well be effective ways to exploit such sparsity and convert it into imaging speed. The Verasonics system we used in [the 2009 IEEE UFFC paper that you mentioned] and in a more recent 2012 IEEE UFFC paper has proven very flexible and programmable, but its 128 channels are far from sufficient to drive the thousands of elements of a 2D transducer as used for volume imaging. On the other hand, clinical 4D systems are not programmable and could not readily be made to emit several simultaneous beams, and they typically are not well focused (on purpose, so that a whole region might be insonified at once). Getting your “reporter devices” to work at pressure levels readily available with a clinical 4D scanner might be easiest as far as imaging is involved.TLD: Thanks for the suggestions. I particularly like the Versatronics strategy of moving more function into the software where they’ll be able to ride on Intel’s coattails — or possibly Nvidia’s GPGPU version of accelerating returns from Moore’s Law. If that sort of functional migration becomes a trend, more hardware manufacturers might be attracted to developing lower cost ultrasound devices and the next thing you know all the gamers will have ultrasound headsets — now there’s an interesting business case for a startup. Thanks again for the feedback and, in particular, thanks mentioning that Benjamin Schwartz has joined Google. I’ll definitely contact him and tell him about our project.
February 21, 2013
I had a discussion this morning with Chi-Yin Lee — an ultrasound engineer, Mike Sekins — Chi-Yin’s manager and the director of Siemens Ultrasound Innovations & Applications, and Shelby Brunke — an engineering manager overseeing several ultrasound projects, all three at Siemens in Seattle. The goal of the meeting was to determine whether and to what extent there are ultrasound technologies that might translate into tools that could be reliably combined to make rapid progress in solving the readout problem. In much the same way that both Ed and Akram have leveraged existing technologies from the semiconductor industry, e.g., semiconductor fabrication facilities, I’m interested mature medical-imaging and materials-testing technologies we can exploit off the shelf. Alas, the story is not nearly as encouraging as it is for CMOS semiconductor and related MEMS fabrication. Here’s what I learned:
It is possible to inject energy into the brain in a highly focused manner either by performing a craniotomy or by using a CT scan to construct an accurate 3-D model of the skull [168, 156] and with this model generate a phased-array scanning algorithm particular to this skull to correct for aberrations in signal propagation due to the variable thickness of the skull. The computations involved are substantial, but they only have to be done once, so that the phased-array wave fronts can be corrected for every location to be scanned in the target tissue. Mike said that this sort of computational modeling has been successfully performed on humans as part of existing therapeutic options.
He told us that micro bubbles are increasingly used in ultrasound as a contrast agent and as targeted agents. Siemens can use a focused beam to create an amplification of the absorbed energy both in terms of heating and mechanical effects such as cavitation. If you focus the ultrasound in an area enclosing a micro bubble you can stimulate a reaction that is “greater than the sum of the parts in a sense because you induce resonance in the micro bubble.
Controlled methods for the delivery of stimuli — excitation of motor cortex neurons causing leg muscles to contract — have been demonstrated but are not off the shelf, require custom devices and are mostly academic demonstrations. There is increasing use of non-thermal stimulation of the brain for applications involving the diagnosis of neurological diseases. There has been a recent “perceived” breakthrough in a new therapy for the treatment of essential tremor. The treatment, currently in trial, employs a focused beam through the cranium using CT-scanned model and corrected phased array deployment algorithm of the sort mentioned above, and, in some cases, has completely eliminated the symptoms due to hyper-stimulation with no deleterious side effects.
I asked Mike and Shelby if Siemens would be interested in partnering with other academic and industrial labs in responding to a BAA for BAM-related research. They’re no stranger to DARPA funding but Mike said this was foo far out of his labs central expertise for him to make a good business case. He did say he would point me to individuals more than happy to work with us on this however. Specifically, somebody who could steer us to resources and is likely to have great personal interest in such a project — a neurologist, brain surgeon and the head of a professional society promoting focused ultrasound for clinical use.
I had high hopes for more technical detail, but none of the engineers present were expert in the underlying physiological issues. Here are some questions I posed to Chi-Yin that he was able to answer:
TLD: What physical units do engineers use to characterize the local changes to tissue resulting from a focused beam scan?
CY: For HIFU, people use MRI for temperature monitoring, the temperature map shows the increase of temperature in Celsius. For diagnostic ultrasound, we use intensity51 (W/cm2).
TLD: How about changes to tissues at equilibrium accounting for repeated scans, unfocused energy, back-scatter, absorption?
CY: The energy delivered to a particular location is defined as: Intensity × Time × Cross Area = Energy. For B mode52 imaging on a commercial scanner, common metrics include spatial peak-temporal average intensity (ISPTA) and mechanical index (MI).
TLD: What about the physical size of existing ultrasound devices given that the early test animals will probably be mice?
CY: The transducer of the ultrasound machine is quite small, usually around a few centimeters depending on whether it is linear, phase or curve. The size of a complete machine can vary widely depending on the application and sophistication of the imaging technology. Nowadays, hand-held ultrasound machines — laptop size — are pretty powerful.
Unfortunately, Chi-Yin was unable to answer my questions relating to the underlying biophysics such as:
TLD: What other changes to cells might we expect, e.g., changes in diffusion rates, ion concentration, protein expression?
TLD: What about tissue density: tables show one number, but brain is composed of fat (~8%), water (~80%), collagen, etc?
Perhaps the neurologist Mike promised to set us up with will be able to point us to some relevant papers. Mike suggested Diagnostic Ultrasound Imaging, Inside Out by Thomas Szabo, Elsevier Press as a good general engineering reference that is reasonably up to day including sections on high-intensity focused ultrasound. I just looked and the most recent edition on Amazon was published in 2004 and the next edition — which you can pre-order — isn’t available for delivery until October 2013. I balked at the somewhat dated material and the more than $400 price tag and am now reviewing Foundations of Biomedical Ultrasound by Richard Cobbold, Oxford University Press, 2007, which apparently covers much of the same material and is less than half the price of Szabo’s text.
One of the engineers at Siemens mentioned BioSono which is a web site that ultrasound engineers working on medical applications frequent. It has a good “Ultrasound Physics” tutorial aimed at engineers with basic, math, physics and signal processing, some useful tissue-parameter tables, simulations and some nice animations for teaching. I also found this article by Haar and Coussios [146] useful though a bit dated. It is interesting that the velocity of sound in the brain as listed in the BioSono tables is given as a single number, 1470 m/s, while that of fat is given as a range, 1410–1479. In other tables, brain tissue and water are listed as having the same velocity. The heart, liver and lung are also ranges but the kidney is again a single number. Seems consistent for heart and lung but I would have expected liver to more similar to the kidney. How much depends on the medium being homogeneous? If the velocity number for the brain is accurate sound must work in bilipid membranes about the same as in cerebospinal fluid. How does the variable thickness of the skull complicate imaging?
On another topic primarily of interest here because of the connection to channelrhodopsins, the company RetroSense Therapeutics has announced a gene therapy that uses stem-cell and recombinant DNA technologies to restore lost function in degenerated photoreceptors by altering the genes in other retinal cells so that they express the photosensitivity gene, channelrhodopsin-2, essentially turning the altered cells into replacement photoreceptors.
February 19, 2013
Here is Akram Sadek responding to some earlier posts from Ed, David and me on ultrasound technologies:
First, with regards Ed’s idea of using microbubbles. I remember many years ago when applying to grad school, meeting Michael Levene at Yale as a prospective. I was very interested in using ultrasound to treat cancer, perhaps by selectively destroying cells based on their diameter. Professor Levene mentioned a fantastic idea he had to use nanoparticles to induce cavitation on ultrasonic irradiation. The whole idea was to get the nanoparticles selectively endocytosed into cancerous cells, which is a difficult research problem in itself. But perhaps a concept like this could be combined with something else Ed mentioned which I had also been thinking about, perhaps relying on the spacing between two nanoparticles. What if these (dielectric) particles sat apart from each other across the neuronal membrane? Their separation might be modulated by the extremely high electric field across it. And then perhaps microbubble formation could be controlled based on the particle separation. You’d use one ultrasonic frequency to induce the bubble formation and another to detect it’s formation.Another point you’ve all raised is the bandwidth problem. As you’ve calculated, the maximum bandwidth in the best case scenario would be 1 million channels. What about trying to solve this problem the way cellular networks do? This is a technique I’m using in my optically-actuated NEMS neural probe. Each cell in a cellular network has a limited bandwidth of frequencies. Instead of trying to impose the same limited bandwidth over the entire country such that each cell phone customer connects via a unique frequency, the bandwidth is limited to cells only a few miles wide. This bandwidth is then repeated in the next cell area. You basically superpose two coding schemes to multiply your bandwidth. Perhaps this could be applied to the ultrasound scheme by combining it with a completely different modality, such as microwave illumination. This way you’d maintain the parallelism of the whole brain “illumination” approach, but now you might have (106)2 = 1012 channels instead. Each of the channels would be addressable, which you would map in space initially using a serial approach as David suggested.
David, to elaborate on my earlier comments about neural probes. In my lab another graduate student (Aditya Rajagopal) is developing neural probes only 250 × 250 × 250μm in size, that are powered optically and that have an on-board low-power microlaser for readout. The chips are essentially CMOS ring-resonators that modulate the output frequency based on the voltage detected by the microelectrodes. They only have a few recording channels per device, but are completely wireless and minimally invasive. Instead of using the laser for the output, you could replace it with a piezoelectric transducer. This is the least elegant way of doing things, converting light into electrical current and then into sound.
What we’re talking about here though is I think the right track, to do it in a more direct fashion through harnessing the physics of interaction between mechanical or electromagnetic radiation and nanoparticles sensitive to ions, electrical or magnetic fields. When I mentioned using piezoelectric NEMS transducers, I meant it in this sense. Your nanodevice would detect the voltage via the piezoelectric effect, which would modulate it’s mechanical properties. This would alter it’s resonance frequency and it’s absorption of ultrasound of a specific frequency. You could detect that absorption externally. So it works like RFID, except using ultrasound and nanoscale particles or devices.
I talked with a couple of Siemens engineers from the Seattle-based ultrasound group about their technologies for high intensity focused ultrasound (HIFU) for therapeutic ablation, parallel scanning for high-resolution real-time cardiac imaging, and phased-array ultrasonics (PAU). They’ve developed some amazing products for industrial materials science and medical applications, including a portable device they developed for DARPA small enough to be carried in the field that can find and cauterize the veins supplying blood to an internal hemorrhage53. It took some time to convey the problem I wanted them to solve. I started with a summary of what I sent around earlier:
TLD: One idea we’ve been thinking about involves using a focused beam — or multiple beams in parallel — to scan the 3-D volume of the target tissue so as to sequentially trigger localized readout. Imagine a scanning beam that can heat a very small volume in such a way as to trigger a nanoscale reporter device to sense its local state and then transmit a coded signal which is received and decoded by a receiver external to the tissue. Since only one location is heated at any point in time only the reporters in that location have to contend for the available frequency spectrum bandwidth.Then we iterated on this to get around their initial effort to pigeonhole the problem into either a HIFU problem with a single focus of attention or a purely imaging problem amenable to their parallel-beam scanning technology. In any case, we seemed to arrive at a mutual understanding. Here’s Chi-Yin Lee’s message and my immediate reply from yesterday evening:
CY: [As I understand it, you need] a very tight beam focus to pinpoint cell-sized targets and a fast scanning technique such as parallel transmission beams to interrogate all the cell-sized targets in a volume. The beam [must] penetrate the skull and dura and deliver the right amount of energy to the target.[With regard to resolution, the] two methods ultrasound engineers commonly use are:
the smaller F — ratio of aperture size over depth — or the larger the aperture size — use more elements, and
the higher frequency, the lower the penetration.
It is a balancing act depending on the situation.
I can see the most challenging part [will be delivering] the right amount of energy to the target — for therapy case, the temperature can rise in tens of degrees, I assume to you need less power in your case.
I will talk with Mike Sekins (our innovation director and a HIFU expert) and some colleagues in ultrasound engineering team about the flexibility of these requirements.
TLD: It occurred to me that it would be very useful know a lower bound on the energy that could be absorbed at a given location without compromising the cells as well as an estimate of how high the temperature might rise in the limit as the tissue is repeatedly scanned at a given energy.
February 18, 2013
The advantage of Ed’s microscopy perspective is that “illumination” of the target cells, whether by light or pressure, happens all at once and so it scales beautifully since every cell or, rather, every transducer responsible for a cell or local collection of cells can then proceed in parallel. The trouble with that approach is that once the transducers have completed their local computations — sensing, coding, etc — they now have to (a) establish cell coordinates to annotate the data they are preparing to transmit and (b) sort out how to share the available bandwidth resources relying on only local information.
Assigning a unique barcode to each cell addresses one aspect of (a) namely reliably identifying each cell with a stream of data, but doesn’t help in determining the relative locations of cells or whether two cells are likely to participate in the same anatomical structure or functional unit. The idea of every cell transmitting data in parallel doesn’t appear to be an option for us due to limited transmitter frequency-spectrum and bandwidth availability and the problem of positioning enough receivers on the skull. Near-field communication technologies could help with respect to the former but I’m guessing not the latter.
The serial-scanning approach doesn’t have near the scaling opportunities afforded by wholesale illumination. However, it is theoretically possible to divide the volume containing the tissue into N × N × N sub volumes so that, assuming N3 non-interfering transmission channels, scanning and readout could be scaled by O(N3). I’m also assuming that the individual ultrasound scanners are small enough that they could be positioned around the skull or tissue sample and configured so that that their signals do not interfere with one another.
The top story at the New York Times this morning: “President Plans Decade-Long Effort to Map Human Brain”. The article leads with “The Obama administration is planning a decade-long scientific effort to examine the workings of the human brain and build a comprehensive map of its activity, seeking to do for the brain what the Human Genome Project did for genetics”. The article makes the obvious analogy to HGP and attempts to disassociate BAM from Markram’s EU-funded Human Brain Project (HBP). They mention Francis Collins who led the NIH HGP effort and apparently leaked the BAM information on his twitter feed, and George Church got his picture posted along with a reference to the recent paper in Neuron that he co-authored with several other prominent neuroscientists [6].
The article gives a nod to big-data and Google, mentioning the Kavli Futures meeting that I participated in back in January: “A meeting held on January 17 at the California Institute of Technology was attended by the three government agencies, as well as neuroscientists, nanoscientists and representatives from Google, Microsoft and Qualcomm. According to a summary of the meeting, it was held to determine whether computing facilities existed to capture and analyze the vast amounts of data that would come from the project. The scientists and technologists concluded that they did.” Well, they exaggerate, but you can’t expect reporters to read, much less comprehend their source material.
February 17, 2013
Ed Boyden suggested54 that photoacoustic imaging might have the necessary resolution and capability to scale to the whole brain we’re looking for, and I agree that something like his acoustic-microscopy idea might work. I’ve been thinking about an alternative strategy using ultrasound to simplify the readout problem. The basic idea is to use focused ultrasound to perform a serial 3-D scan of the tissue. Nanoscale detectors distributed through the tissue would be triggered by the ultrasound scan to sense cell-membrane potentials or detect the presence of target molecules. The resulting sensor data would then be encoded and transmitted to an external receiver. In the simplest scenario, all of the transmitters could use the same channel and transmit to a single receiver.
In a more sophisticated setting, the ultrasound scanning could serve multiple purposes: (a) to serialize readout so as to optimize available bandwidth — in the case of a completely serialized scan, only one location transmits at a time, (b) to precisely target locations for excitation or inhibition, (c) to provide energy using a piezoelectric transducer to transmit the local sensor output to an external receiver. Scanning need not be done in completely serial order; one could use several beams in parallel as long as the beam foci are separated spatially [100] to mitigate transmission interference. We also get a precise location for the sensed cell activity for free. The biggest challenge would be to build a nanoscale package capable of recognizing the scan signal, encoding the local state and transmitting it beyond the skull. The packages would be free floating since they just report on the state of their current location wherever that might be.
This approach has the advantage that applications in medical imaging are accelerating the development of ultrasound technology for rapidly scanning tissue at high temporal and spatial resolution — the physician really wants to see in real-time the blood flow through that mitral valve or the restriction in that coronary artery. Increasingly these devices are handheld so examinations can be conducted at bedside or in a primary-care facility, thus imposing additional challenges for commercial applications that we could exploit. Many of the tricks in the optical engineers toolkit are being applied to speed scanning — see for example this paper demonstrating order-of-magnitude improvements by using multiple beams at once: Accelerated Focused Ultrasound Imaging. Bruno Madore, P. Jason White, Kai Thomenius, and Gregory T. Clement. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. Volume 56, Number 12 (2009).
Similarly improvements in the accuracy and power of the focused beam are being driven by the therapeutic benefits of ultrasound [10] ablation as a an alternative to more invasive methods such as using a Gamma Knife or conventional surgery — see for example: Carbon-nanotube optoacoustic lens for focused ultrasound generation and high-precision targeted therapy. Hyoung Won Baac, Jong G. Ok, Adam Maxwell, Kyu-Tae Lee, Yu-Chih Chen, A. John Hart, Zhen Xu, Euisik Yoon, and L. Jay Guo. Scientific Reports, Volume 2, Number 989, Pages 1-8 (2012).
February 15, 2013
Here is a selection of BAM-related papers referenced in a recent message from Akram Sadek, a graduate student at Caltech working in the lab of Axel Scherer, who did the cochlea-inspired nanoscale multiplexer based on piezoelectric nanomechanical resonators [133]. Actually, the penultimate reference is not to a paper but to a talk by Shimon Weiss from UCLA and the last reference is to a project description found on the web site for Mark Schnitzer’s lab at Stanford:
Zhong Lin Zhang and his lab at Georgia Tech pioneered in the development of piezoelectric and nanoscale-cantilever-beam-based nanogenerators. Here they demonstrate the a self-powered system driven by a nanogenerator that works wirelessly and independently for long-distance data transmission. — Self-Powered System with Wireless Data Transmission. Youfan Hu and Yan Zhang and Chen Xu and Long Lin and Robert L. Snyder and Zhong Lin Wang. Nano Letters Volume 11, Number 6, Pages 2572-2577 (2011). (HTML)
This is an interesting follow-up to an earlier paper [7] on the ephatic coupling of cortical neurons by Christof Koch, Henry Markram and their collaborators. — The origin of extracellular fields and currents — EEG, ECoG, LFP and spikes. György Buzsáki and Costas A. Anastassiou and Christof Koch. Nature Reviews Neuroscience. Volume 13, Pages 408-420 (2012).
Here is an early paper addressing the BAM challenge in which the author opines “[a]chieving the full potential of massively parallel neuronal recording will require further development of the neuron-electrode interface, automated and efficient spike-sorting algorithms for effective isolation and identification of single neurons, and new mathematical insights for the analysis of network properties. — Large-scale recording of neuronal ensembles. György Buzáki. Nature Neuroscience. Volume 7, Number 5, Pages 446-451 (2004).
The review article begins by claiming that “[e]ffective non-invasive control of neural activity can be achieved with optically excitable quantum dots, tiny semiconductor particles nanometers across”. — Quantum dot optical switches for remote control of neurons. Lih Lin. SPIE. 30 April 2012. (HTML)
The authors “propose a new way to make cells photosensitive without using genetic or chemical manipulation, which alters natural cells, in conjunction with quantum dots”. — Remote switching of cellular activity and cell signaling using light in conjunction with quantum dots. Katherine Lugo and Xiaoyu Miao and Fred Rieke and Lih Y. Lin. Biomedical Optics Express. Volume 3, Number 3, Pages 447-454 (2012). (HTML)
In this review article, the authors “cover different voltage imaging methods (including organic fluorophores, SHG chromophores, genetic indicators, hybrid, nanoparticles, and intrinsic approaches) and illustrate some of their applications to neuronal biophysics and mammalian circuit analysis”. — Imaging Voltage in Neurons. Darcy S. Peterka and Hiroto Takahashi and Rafael Yuste. Neuron. Volume 69, Pages 9-21 (2011).
This solid but now somewhat dated introduction to optical methods leads by stating “Electrophysiology, the ’gold standard’ for investigating neuronal signalling, is being challenged by a new generation of optical probes. Together with new forms of microscopy, these probes allow us to measure and control neuronal signals with spatial resolution and genetic specificity that already greatly surpass those of electrophysiology. We predict that the photon will progressively replace the electron for probing neuronal function, particularly for targeted stimulation and silencing of neuronal populations”. — Electrophysiology in the age of light. Massimo Scanziani and Michael Häusser. Nature. Volume 461, Number 15, Pages 930-939 (2009).
This paper is also dated but interesting for historical purposes: “Nanotechnologies exploit materials and devices with a functional organization that has been engineered at the nanometre scale. The application of nanotechnology in cell biology and physiology enables targeted interactions at a fundamental molecular level. In neuroscience, this entails specific interactions with neurons and glial cells. Examples of current research include technologies that are designed to better interact with neural cells, advanced molecular imaging technologies, materials and hybrid molecules used in neural regeneration, neuroprotection, and targeted delivery of drugs and small molecules across the blood-brain barrier:. — Neuroscience nanotechnology: progress, opportunities and challenges. Gabriel Silva. Nature Neuroscience. Volume 7, Pages 65-73 (2006).
By transmitting [ultrasound] waveforms through hippocampal slice cultures and ex vivo mouse brains, [the authors] determined low-intensity, low-frequency ultrasound is capable of remotely and noninvasively exciting neurons and network activity. — Remote Excitation of Neuronal Circuits Using Low-Intensity, Low-Frequency Ultrasound. William J. Tyler and Yusuf Tufail and Michael Finsterwald and Monica L. Tauchmann and Emily J. Olson and Cassondra Majestic. PLoS ONE, Volume 3, Number 10. Pages e3511 (2008). (HTML)
William Tyler is a co-founder of Neurotrek based in Silicon Valley and Roanoke, VA. Neurotrek is commercializing the use of ultrasound technologies and devices for noninvasive brain stimulation. Here are some additional papers on their web site:
Noninvasive functional neurosurgery using ultrasound. William J. Tyler and Yusuf Tufail and Sandipan Pati. Nature Reviews Neurology Volume 6, Pages 13-14 (2010). (PDF)
Transcranial Pulsed Ultrasound Stimulates Intact Brain Circuits. Yusuf Tufail and Alexei Matyushov and Nathan Baldwin and Monica L. Tauchmann and Joseph Georges and Anna Yoshihiro and Stephen I. Helms Tillery and William J. Tyler. Neuron Volume 66, Pages 681-694, (2010). (PDF)
Ultrasonic neuromodulation by brain stimulation with transcranial ultrasound. Yusuf Tufail and Anna Yoshihiro and Sandipan Pati and Monica M Li and William J Tyler. Nature Protocols Volume 6, Number 9, Pages 1453-1470 (2011). (PDF)
Super-resolution optical microscopy is a rapidly evolving area of fluorescence microscopy [...] impacting many fields of science. [Shimon Weiss] presents a novel Super-resolution method that affords a favorable trade-off between speed and resolution and therefore is well suited for following cellular processes in live cells. He will also report on our efforts to develop quantum-confined Stark effect-based nanovoltage sensors for all-optical membrane potential recording — Superresolved Ultrasensitive Optical Imaging and Spectroscopy. Shimon Weiss. (HTML)
Development of high-throughput, massively parallel imaging techniques for studying brain function in large numbers of Drosophila concurrently. [...] We are constructing instrumentation allowing the brain volumes of ~100 alert flies to be imaged simultaneously by two-photon fluorescence microscopy. — This is just something I ran across when looking at Mark Schnitzer’s lab pages at Stanford. Previously I discovered his papers on molecular motors and found them informative. There appear to be several interesting papers coming out of this lab recently. (HTML)
Here’s what Akram had to say about Tyler and ultrasound methods:
I remember a few years back listening to a lecture by William Tyler on his work using ultrasound for neural stimulation. This technique stimulates relatively large volumes of brain tissue, so it isn’t very specific, but it is of course non-invasive. During his lecture, Professor Tyler mentioned he was planning on developing mechano-sensitive ion channels that would be specific for the ultrasound. This would in principle enable single neuron specificity.Recording though is a very different problem. If you use ultrasound in the MHz range, there should be sufficient bandwidth for hundreds of neural channels. In principle, I think it could certainly be used for recording somehow. The problem though is the absorption coefficient for ultrasound. For brain it is about 0.6 dB / (MHz cm). For bone though it is an order of magnitude greater. And of course, the greater the frequency the stronger the tissue absorption. This may not be as bad a problem as it seems though. Even if there’s strong absorption and reflection at the interface between the brain, skull and skin, you can make your external detector very sensitive, as size is not really an issue. Depending on the power of your implant though, the SNR might be too low. I’d have to do a back of the envelope calculation to get an idea.
Simplest way to record would be to adapt a neural probe we’re developing to use a piezo transducer instead of a laser for the data output. I suspect though the power consumption would be too high for sufficient SNR at the receiver. Another possibility is using piezo NEMS or transducers to directly detect the neuron voltage, and then use external ultrasound and measure how the absorption is modulated by the devices. Again, it’s unclear how poor the SNR would be. But certainly it’s a possibility I think. In terms of powering an implantable device with ultrasound, some really interesting work has been done on this using piezoelectric zinc-oxide (ZnO) nano wires, by Zhong Wang at Georgia Tech. His device also transmits wirelessly, but they use RF to do so and used the ZnO nano wires to also construct a super capacitor to build up enough voltage to power everything.
Related to Akram’s comments above, here is an interesting review of Lihong Wang’s 2011 plenary talk at the Physics of Quantum Electronics Conference on photoacoustic imaging. In this technology, the target tissue is hit with a pulse of light that thermally induces presure — differentially depending on the local density of the tissue — producing ultrasonic waves that are picked up with a microphone. The question of resolution is somewhat complicated but intriguing, and this short review article [162] in Science is reasonably concise in explaining the issues55.
February 13, 2013
Here are some basic facts and figures about radio transmission that I anticipate will prove useful to students when we get around to considering how to engineer communication networks to readout neural-state information:
The electromagnetic (EM) spectrum spans relatively slow radio waves (50-1000 MHz or wavelengths from 10m to somewhat less than a meter) through microwaves and infrared (IR) to the visible (700-400nm) and then through ultraviolet (UV) to X-rays and Gamma rays (0.01nm). Electromagnetic waves travel at the speed of light in a vacuum, but travel slower when traveling through a medium such as water or glass. According to the wave equation v = f λ where f is the frequency and λ is the wavelength we have v = c in the case of a wave traveling through a vacuum where c = 3.0 108 is the speed of light in a vacuum. Their frequencies do not change when they travel from one medium to another, e.g., from water to glass, because their frequency depends only on the source of the wave. Only their speeds and wavelengths change from one medium to another.
Electromagnetic waves can be classified as either ionising or non-ionising56. Ionising radiations are extremely high frequency waves including X-rays and gamma rays. In general, radiation with energies in the ultraviolet (UV) band or higher is ionising. Radio waves are not ionising — the only known biological effect they have on the human body is heating. UV radiation — 10-8m to 10-7m — cannot penetrate tissue but can cause superficial damage including genetic mutation leading to skin cancer. X-rays — 10-13m to 10-8m — and Gamma rays — 10-14m to 10-10m — do penetrate tissue and can cause cellular damage deep within tissue.
The amount of RF energy that is absorbed (deposited into) human body tissue and the rate at which the energy decreases with the depth of penetration depends on both the type of tissue57 that the energy passes through and on the frequency of the incident radiation. At radio frequencies below 4 MHz the body is essentially transparent to the energy. As the frequency is increased more energy is absorbed by the human body. At high frequencies — around 1 GHz (1000 MHz) or more — there is scattering. Increasing the frequency of electromagnetic energy still further, the point is reached when the body reflects radiation (visible light). The refractive index n of an optical medium like water is defined as n = c / v where c is the speed of light in a vacuum and v is the speed of light in the medium. The refractive index of air is 1.000297 and that of water is 1.333. The brain is 75-80% water and so we’ll approximate its refractive index to be the same as water58. Since v = f λ we have c / n = v λ and so the wavelength of a signal entering the brain will decrease by a factor of approximately 1 / 1.33 = 0.75. Much of this also applies to acoustic signals for neural recording and stimuation and you might want to check out Akram Sadek’s back-of-the-envelope musings here.
For the most part we can assume that electromagnetic waves of different frequencies combine linearly. This provides ample opportunity for engineers to employ multiple transmitter frequencies that can be separated at the receiver end of the transmission. Modern mobile telephone technologies combine techniques relying on multiple transmission frequencies and sending multiplex multiple signals — data and phone conversations in the case of cell phones — using each individual frequency. These technologies include Frequency-Division Multiplexing (FDM), Time Division Multiple Access (TDMA) and Code Division Multiple Access (CDMA). Check out Chris Uhlik’s back-of-the-envelope calculations for a multiple-receiver and multiple-transmitter solutions here.
Near-field communication (NCF) — mentioned in Chris Uhlik’s comments — involves communication in which the transmitter and receiver are in close proximity, usually no more than a few centimeters59. NFC technologies are one approach to Radio Frequency Identification tagging (RFID). Near-field methods are of particular interest for communications in brain tissue due to the fact that that the components in the brain can be powered by an external power source using the same components required for communication. NFC range is limited by the extent of the near field which is determined by frequency with higher frequencies having smaller effective near fields and with lower frequencies we have lower data transmission rates. The general rule of thumb is that the near field extends one or two wavelengths from its source. Frequencies greater than 300 MHz (microwave) are generally considered far field while those less than 300 MHz are considered near field [68].
Think about adding a section on the relationship between transmission frequency and the minimum size of the transmitting and receiving antennae: see the following pages on radio antennae, antennae and electrical length, and the formulae for calculating antenna length versus frequency.
February 11, 2013
As the field of neuroscience seeks to justify increased public funding, the focus has been on relatively rare disorders like locked in syndrome and Parkinson’s disease or relatively common disorders primarily targeting the older population like dementia and Alzheimer’s disease. While important, this emphasis neglects a board class of common disorders relating to mood and sleep regulation that have a huge economic and social impact. These disorders involve the dopamine reward centers implicated in substance addiction and compulsive behavior, diffuse rhythmic signaling pathways important in sleep regulation and memory consolidation, and circuits involving the amygdala and hippocampus thought to play a role in anxiety and depression. These disorders tend to be difficult to diagnose and treat, often have associated social stigmata, can occur at any age, and manifest both recurrent and chronic variations. They have a substantial economic impact in terms of lost productivity due to poor or insufficient sleep, depressed or exaggerated mood and common cognitive deficits associated with anxiety, stress, poor diet, and self-medication involving alcohol, caffeine, other over-the-counter and prescribed mood and attention-altering substances.
While it is difficult to quantify the full economic impact due to the interdependence of symptoms, there have been a number of studies that focus on individual contributing factors. A 2003 article in JAMA entitled “Cost of lost productive work time among US workers with depression” states that “Excess lost productive time (LPT) costs from depression were derived as the difference in LPT among individuals with depression minus the expected LPT in the absence of depression projected to the US workforce” and estimates that “US workers with depression cost employers an estimated 44 billion dollars per year in LPT, an excess of 31 billion dollars per year compared with peers without depression. This estimate does not include labor costs associated with short- and long-term disability.” A 2006 report issued by the Institute of Medicine (US) Committee on Sleep Medicine and Research entitled “Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem” states that although “problems falling asleep or daytime sleepiness affect 35 to 40 percent of the population [Hossain and Shapiro, 2002], the full economic impact of sleep loss and sleep disorders on individuals and society is not known.” A Harvard Business Review article entitled “Sleep Deprivation’s True Workplace Costs” is not so timid and reports that across four companies studied “sleep-related reductions in productivity cost $54 million a year. This doesn’t include the cost of absenteeism–those with insomnia missed work an extra five days a year compared to good sleepers.”
What is holding up progress? Despite the so-called decade of the brain (1990-2000) neuroscientists have by and large failed to take advantage of advances in sensing and computation. A number of us have been working to promote scalable neuroscience and develop new funding initiatives [6] to support promising research in this area. Another impediment to progress which is compounded by the slow pace of the field concerns the fact that, while excitement in the 90s and early 00s attracted top talent to the best graduate schools, many of those students ended up disappointed and chose other fields following graduation. We have at Google several accomplished neuroscientists productively employed on revenue-producing projects, passionate about neuroscience, but discouraged with the pace of the field in which they did their doctoral research.
Recently several of us have been working to identify both near- and intermediate-term projects whose enabling technologies are fueled by advances in neuroscience, advances in other disciplines such as nanotechnology that would accelerate neuroscience research, and opportunities to work with or fund scientists whom we think are on the cusp of break-through technologies. In terms of shorter term opportunities we have been looking at how to leverage cell phone technology to deliver on insights derived from the brain sciences. We have contacts in several labs — including University of Washington and Johns Hopkins — working on retinal implants and prosthetic devices for the visually impaired. We have also been talking with scientists at New York University Medical School about non-invasive diagnostic tools for brain trauma that can be built from simple off-the-shelf technologies — basically eye-tracking and computational resources of Android phone or tablet, which, in several surprising cases, performs better than expert diagnosticians and detects abnormalities60, e.g., concussions, that more expensive, invasive and complicated-to-administer procedures such as MRI and PET scans miss. Moreover, the necessary hardware is available and more powerful related applications could be implemented using HUD technology based on Google Glass.
One application we are currently looking at involves developing a true sleep manager in contrast to Zeo’s version which is really a sleep record keeper that may or may not prove useful in managing your sleep. Relevant technologies include EEG for monitoring brain rhythms, near-infrared sensing, heart rate, skin conduction, and muscle electrical activity for arousal markers, tDCS, TMS, facial heating and ambient light and sound for non-invasive interventions. Recent research has shown that it is possible to speed the onset of sleep, modify the duration of its multiple stages and even induce sleep so as to shorten the time require for a full, restorative sleep by as much as 25%. The economic impact of such a technology would be enormous. See the recent article by Stickgold and Walker [127] for a good review of what’s known about sleep and memory.
Ditto for a true mood manager that would benefit from new sensor technologies including blood-sugar, facial cues, eye tracking, and more-accurate EEG with multiple channels reading from diverse sites on the skull. There is already a growing market for cell phone apps that provide various tools for tracking and analyzing mood:
Moodlytics provides a generic mood tracker with analytics, goal setting and data backup.
eMood provides a free mood tracker Android app for people with bipolar disorder that also keeps track of medications and sends summary reports to your doctor if you so wish.
Radiant Monkey Software provides event trackers tailored to depression, anxiety and other disorders.
But there is a great deal of potential yet to be realized and the key will be to improve wearable sensor nets to track markers relating to mood and ultimately complete the loop using non-invasive interventions such as transcranial magnetic stimulation which have already shown dramatic results for drug-resistant depression and various developmental and geriatric applications. Indeed it might be argued that neuroscience is at a stage comparable to particle physics in the late 1800s with too many theories chasing too little data.
As for scalable neuroscience and accelerating the pace of research in the brain sciences, we are not satisfied with progress. The Human Brain Project (HBP) is, in our opinion, a colossal waste of money for the EU; they’ve been duped into believing the brain can be cracked like the Higgs Boson. The difference in the maturity of the parent disciplines, neuroscience and particle physics, is considerable. The Brain Activity Mapping (BAM) Project modeled after the Human Genome Project is more likely to make progress but vested interests and the process governing federal funding is likely to be wasteful. What can we do? In addition to the short term project sketched above, we intend to continue our partnerships with the Church (Harvard), Boyden (MIT), Kopell (Boston University), Smith (Stanford) and Olshausen (Berkeley) labs and others who understand the importance of automation and scaling and are working on key enabling technologies. We have also started to build relationships with some of top private institutions like Allen Institute for Brain Science. We are also interested in working with bio- and neuro-savvy partners at Google Ventures like Bill Maris and Krishna Yeshwant to identify relevant projects, incubate promising new startups, and provide capital to existing companies that building better sensing and imaging technologies.
We are particularly interested in labs populated by multi-disciplinary teams of scientists and engineers, building instruments and developing new processes, automating their experimental procedures and, critically, sharing both the data they produce and their hardware designs. Several key enabling-technology efforts have been open-sourced, e.g., optogenetics and synthetic biology, electroencephalography, and it is becoming more and more common as researchers discover the advantages of open collaboration. We already have contacts at the Allen Institute for Brain Science, Cold Spring Harbor Laboratory and Salk Institute Computational Neurobiology Laboratory and are interested in pursuing collaboration with Janelia Farm Research Campus of HHMI. We are cautiously optimistic about the prospects for substantial progress over the next few years and one dimension of our ongoing effort is to make sure that Google is aware of new developments, participating where it makes sense, and looking out for opportunities to accelerate research.
February 9, 2013
Scattered notes from a meeting to discuss photoacoustic neural imaging and intervention technologies with Ed Boyden61, Greg Corrado, David Heckerman and Jon Shlens on February 8, 2013, at Google Labs in Los Angeles:
Surgically introducing ultrasound transducers through nasal or oral cavities or by exploiting variability in cranial thickness62.
Using 3-D probes developed in Ed’s lab for calcium imaging by outfitting fiber-optic channels with nanoscale photosensors.
Canadian group working on “red-white-spectrum” calcium imaging; also infrared optogenetic technologies lagging green63.
Control changes in the shape of microbubbles to alter their light or acoustic signal scattering characteristics.
Engineer microbubbles so that it takes very little energy to institute a phase transition in the bubble contents.64.
New work on calcium imaging at Janelia Farm capable of single action potential accuracy if spikes are sparse65.
There was an article on sleep in the New Scientist by Jessa Gamble [58] whose TED talk on how to sleep went viral a couple of years back. In this article, Gamble talks about various technologies used to track, modulate and even induce sleep. These technologies include a device used by the military to enable soldiers in the field to catch a quick nap when under prolonged stress. The portable device utilizes facial warming to speed the onset of Stage 1 sleep and an EEG and blue light to wake the subject gradually so as to avoid confusion and disorientation.
Gamble also reports on a procedure involving transcranial Direct Current Stimulation (tDCS) developed at the University of Lubeck, Germany which allows researchers to shift a sleeping subject between adjacent sleep stages with no ill effects. The research described in this article by Marshall et al [104] requires that the subject is already asleep in order effect such shifts.
In another paper by Massimini et al [107] featured in Gamble’s article, the authors describe a method developed by a team of researchers at the University of Wisconsin-Madison led by Giulio Tonni based on Transcranial Magnetic Stimulation (TMS). Unlike the tDCS method this one is capable of inducing sleep by using magnetic stimulation strong enough to trigger a deep sleep and even move a subject directly from sleep onset to Stage 3 bypassing Stages 1 and 2 altogether.
The authors claim that this should enable a person for whom the optimal sleep interval is 8 hours a day to reduce this to 6 hours a day thereby reclaiming 2 hours a day for productive work. This would add a few weeks to the average person’s year, thus effectively lengthening their lives. It’s interesting to think about getting more out of life by simply sleeping less.
February 7, 2013
Draft prologue for a possible CS379C “meta” lecture on the politics and “organizational mismanagement” of modern science and technology:
Science and technology are slow to integrate new information and new ideas. Academic and corporate advancement for scientists and engineers is hampered by counterproductive incentive structures. The “drunk under the lamppost” cartoon and the old adage “science advances one funeral at a time” — a pithy paraphrase of a longer comment in German by Max Planck which I heard from John Tooby. Why should you care? One reason is that your life and that of your children — in terms of span of years, health and physical well being, security and opportunity for self realization — will be much the same as your parents unless your generation changes the status quo. It might be argued that you bear a larger responsibility than the average person based on your advantages in education and opportunities for advancement. Despite a growing global population, the number of highly intelligent, well educated, articulate, ambitious and entrepreneurial people remains relatively small and the barriers to progress that need to be overcome remain high.
We squander our human capital by pitting one scientist against another, encouraging the cult of the individual and discouraging sharing and teamwork, allowing individuals to dominate in directing their research, stressing the positives of self-reliance and personal investment and ambition while largely ignoring the negative consequences of human decision making such as our focus on anecdotes, problems in assessing and effectively taking into account risk and probability, difficulty absorbing spent cost and estimating opportunity cost, and the degree to which losses unduly influence behavior. There is much to be appreciated about human cognition but most would agree it is far from optimal and, with the right sort of leadership, could be considerably improved upon.
Beware of vested interests, you’ll find them lurking behind most decisions. The trick is to learn to distinguish warranted investments in good people and good ideas from self-serving adherence to the status quo that ignores new evidence and an evolving context. And don’t believe that status quo thinking is the unique province of the older, richer or more established segments of the population. In seeking advice on what to work on or how to guide your career, learn to listen and ask the right questions. Instead of asking an accomplished scientist what he or she believes is the most important result or the most important problem to focus on, ask them to tell you about the second and third most important, or better yet, ask them about the craziest idea they’ve heard lately from an otherwise credible source or wunderkind graduate or postdoctoral student. Try to ferret out what they believe is the most disruptive idea threatening the foundations of their discipline. See if you can get them to reveal some conjecture or body of evidence that threatens their one of the pet theories. Get them to talk for awhile and listen carefully.
February 5, 2013
There was an article in the latest issue of the Economist about the race to invent a battery technology that can compete with fossil fuels. The article had some interesting things to say about work at Argonne National Labs, but what caught my eye was mention of a “rapidly growing encyclopedia of substances created by Gerbrand Ceder of the Massachusetts Institute of Technology. Dr Ceder runs the Materials Project, which aims to be the ‘Google of material properties’. It allows researchers to speed up the way they search for things with specific properties.” The Cedar Lab has a bunch of resources relating to this effort66 which goes by the name of the Materials Project.
In this same issue there was an article on remembering and forgetting. Mostly it was full of discouraging news about how memory degrades as we get older and how we fare even worse following sleep — which is likely to be shorter and shallower in older people. I try to meditate for half an hour right before I go to bed and that helps me to get to sleep. I also engage in a form of lucid dreaming in which I recall things I’ve learned during the day and want to remember, or review a problem that I would like to solve. The Economist article prompted my curiosity about whether and to what degree one could engage in active forgetting — a form of lucid erasure as it were — using the meditative practice of allowing thoughts to surface but not paying them close attention or merely noting their appearance. Perhaps you could modify this practice slightly by noting only that you wish to recall or forget certain thoughts. Somehow you would have tag these memories so that the appropriate action — recall or forget — is initiated during sleep. A quick search revealed that Nietzsche discusses active forgetting in his “On the Uses and Disadvantages of History for Life” discourse.
Another quote from Rosenberg and Birdzell [129] that might provide the fodder for a thought exercise to understand the role of networks in scalable commerce:
The first four centuries of the period of mercantile expansion coincided with the period of greatest growth and development on feudal society — an apparent anomaly made possible by the peculiarly feudal and pluralistic device of chartering, outside feudal jurisdiction, towns where trade could prosper. In the simplest case, the expansion was a response to the pressures of what moder economists can recognize as comparative advantage, the advantage of regional specialization inside and outside Europe. Especially after the collapse of military feudalism and the emergence of centralized monarchies, the response to comparative advantage, was untidy — compounded by piracy, smuggling and political corruption as well as of industry, diligence and thrift. It interacted with technological development, each fueling the other. In this period of expansion, Western Europe created an active merchant class and marked out an arena where it could trade with enough freedom to leave it a world of opportunities read to hand. It created also a network of markets, commercialization and financial relationship and economic institutions which needed only to be scaled upward in order to process a much larger volume of trade; no substantial generic modifications were necessary for them to exploit the technology of the Industrial Revolution. — Page 109 [129].
February 3, 2013
BAM is definitely a “big-science” project with lots of savvy players pursuing vested interests, public and private institutions like NIH, Harvard, MIT, Salk and Allen jockeying for a stake in resources that might become available, large potential payoffs in terms of public health, basic science and commercial applications, as well national pride and international competition. It is also a project in which systems neuroscience plays a role front and center and the technologies involved span an incredible range of disciplines. In one respect, the European Commission in funding HBP is right in that such an ambitious program needs scientific oversight and some degree of central authority and planning. My concern is that they are applying antiquated economic policy; they are essentially granting a monopoly to a small group of scientists and in so doing discouraging entrepreneurs from participating and ignoring the power of capital markets to identify and bankroll promising players. Hopefully the United States, if it decides to support BAM, will let the markets pick winners and losers and depend more on individuals to self-organize to create scientific products of broad value to a diverse community of contributing scientists working on BAM-related projects.
Here are the invited speakers for Ed Boyden’s Neurotechnology Ventures course at MIT for Fall 2012:
Speaker: Vinay Gidwaney — DailyFeats
Title: Neurotechnologies for improving behavior and decision-making
Speaker: Ariel Garten, CEO — InterAxon
Title: Thought controlled computing: from popular imagination to popular product
Speaker: Ben Rubin — Zeo
Title: Zeo: a sleep analysis company
Speaker: Casey Stengel — Neuralynx
Title: Neuralynx: an electrophysiology tools company
Speaker: Vincent Pieribone — Marinus
Title: Marinus Pharmaceuticals: a neurosteroid development story
This is an odd bit of background research relating to a lecture I’m planning for my Stanford class that provides an introduction to the role of capital markets, technology and basic science in modern economic theory. I wasn’t much interested in economics and finance until I read William Bernstein’s [17] The Birth of Plenty which hypothesized that the origins of global prosperity were to be found in the development of property rights (the motivation for improvement fueled by ownership), the rule of law (the foundation of stable commerce), capital markets (the democratization of entrepreneurial pursuit) and scientific rationalism (the internalization of the scientific method and the parallel acceleration of technology) — my parentheticals. I was hooked and immediately had to read Adam Smith and a host of other economists and financial theorists and historians. Bernstein’s [18] A Splendid Exchange — which I was quick to notice had the same number of characters (but not letters) as his earlier book — was equally eye-opening for its globe spanning treatment of trade. My latest find is Rosenberg and Birdzell’s [129] How the West Grew Rich: The Economic Transformation of the Industrial World and, while more academic in its content and more pedantic in its prose when compared to Bernstein’s lively writing, I found lots of historical insights relating to capital markets and technology.
Given the interest in China as a growing superpower and economic juggernaut, I was particularly interested in the authors’ discussion of the “recent” history of China — primarily the last two millenia — and why China was relatively late to the game even ignoring the twentieth century and their dalliance with communism. They carefully dissected Joseph Needham’s argument for why China didn’t experience the exponential explosion in wealth that much of western world did while having achieved a comparable starting point in terms of science and technology. The telling contrast was between the west being dominated by a ruling class entrenched by hereditary succession and China ruled by mandarins — the English word is derived from Portuguese and means “to think”, a class of civil servants selected on the basis of their merit with no prospect of hereditary succession. What a wonderfully rational approach to ruling a nation! Needham’s conclusion is that the social and cultural values of Asian “bureaucratic feudalism” were simply incompatible with capitalism and, for that matter, modern science — source Chapter 3, Page 87 of [129].
Despite the seeming egalitarianism of their “rise to power by academic examination” approach to electing the ruling class, and their appreciation of learning in general and scientific knowledge and its application to technology for human benefit in particular, China failed to capitalize (literally) on the rational foundation of the civilization. Europe was, in an important sense, favored by the inevitable decay of its inefficient variant of feudalism which left an opening for the rise of a predominantly mercantile society. Needham points out that the reason was that “the mandaranite was opposed not only to the principles of hereditary aristocratic feudalism but also to the value-systems of the wealthy merchants” — capital accumulation was not open to the scholar-bureaucrats who ruled China but they were not required to pursue an ascetic life-style and few denied themselves pleasure. They had better technology — gunpowder, ships capable of carrying heavy cargo on long voyages, etc — but were contemptuous of material goals and hostile to bourgeois values in general. One of the technologies in which they excelled, namely accounting, was conspicuously absent in medieval Europe and a requisite for developing an effective mercantile technology.
Rosenberg and Birdzell also did a good job of presenting and arguing the merits of Marx and Engels’ theory that colonialism and trade expansion fueled the industrial revolution which in turn caused the explosion in science and technology rather than the other way around. Ideologically Marx and Hegel were opposed to a system that involved “extracting the largest possible ‘surplus value’ from the labor force, but dismissed the value of how markets and competitive pressures compelled capitalists to plow their profits into the expansion” of the very system that creates opportunities for individuals of all classes to achieve a standard of living in keeping with their energy and ambition. Rosenberg and Birdzell conclude that while there may have been periods in which the trends would appear to support this hypothesis, a broader view of history makes it difficult to establish cause and effect in this case and industrialization, technology development and scientific advancement could best be seen as synergistic, each fueling the other two. They do make the claim in Chapter 5 that there are grounds for believing that “the technology of industrial revolution ignited a nineteenth-century growth of markets which dwarfed the earlier expansion.”
February 1, 2013
Note to Terry Sejnowski following a talk at Google by Mike Hawrylycz of the Allen Institute for Brain Science. Mike and I had a short but intense discussion after his talk and promised to VC next week to follow up on many points of shared interest:
Over the last couple of weeks, I had the opportunity to talk with Kevin Briggman — formerly of Winfried Denk’s lab and now starting his own lab at NIH — and Mike Hawrylycz who heads up the informatics team at Allen Institute and is responsible for most of the computing infrastructure behind the Mouse Brain Atlas project. Mike was employee #5 and now along with Clay Reid reports directly to Christof. For all that I enjoyed Clay’s input and appreciated his perspective at the Kavli meeting, Mike is the big-data person at Allen and has a wealth of useful practical knowledge about ontologies, registering neural tissue samples against standardized anatomical reference maps, scaling data collection, building tools for exploring cross correlated datasets and a host of other problems that we talked about at Kavli.
His description of how they collected, collated and built analytics for the Mouse Brain Atlas was extensive and detailed as was his extrapolation to how we might scale up to a human brain atlas. It was considerably easier for them working with the mouse; in particular developing a reference atlas was easier since they worked working with a strain of lab mice — C57BLACK6 — which they raised under near identical conditions and sacrificed at the same exact age and even the same time of day to control for diurnal variation. These mice are inbred and exhibit very little genetic variation. Mike outlined the registration process — necessary since they used many of these nearly identical mince to generate the atlas. He also highlighted the challenges of using machine learning to segment cell body — Briggman had an even more nuanced perspective on this aspect of scaling serial-section microscopy to larger tissue samples.
The Allen Institute is now working on seven different variants at different points during development, each with a different reference atlas. In addition to the genomic / expression maps they’ve also created a connectome atlas — essentially a Van Essen-style connectivity map but in three dimensions — with the ability to do gene expression overlays. The Anatomical Gene Expression database supports powerful searches for correlations among genes and expression patterns — they discovered that while reference-registered brain areas are nice to have for some purposes a detailed 3-D volumetric model with fine-grained voxels is much more useful to scientists. Needless to say that have very sophisticated data collection, storage, annotation and search technology. He also described their high-throughput fluorescence-imaging technology which is quite impressive.
While watching Kevin Briggman’s talk I used his estimates for SEM-voxel size and mouse-brain volume to come up with the following data storage estimate: One voxel from a serial-block-face scanning electron microscope (SBFSEM) of the sort used in Denk’s lab is 10 × 10 × 25 nm or 102 * 25 = 2,500 nm3. The volume of a mouse brain is about 103 mm3. A millimeter is a million nanometers, a cubic millimeter is a million-cubed cubic nanometers and so a mouse brain is (10 * 106)3 = 1021 nm3 and a complete scan would span (1021 / 2,500) SBFSEM voxels. A terabyte is 1,099,511,627,776 bytes and so the number of terabyte disks we would need to store a complete scan is (1021 / 2,500) / 1,099,511,627,776) or approximately (1021 / 2,500) / 1012 = 400,000 TB disks and I suspect I’ve grossly underestimated.
They’ve done all the back-of-the-envelope calculations we have and more. Mike said that the cost of the mouse atlas is in the 10s of millions, and a reasonably straightforward extrapolation to humans would cost 100s of billions, but they have lots of ideas about how to reduce cost and accelerate data collection. We also talked at some length about JPEG compression issues — Mike was visiting Google both to seek out collaborators and to learn from our engineers about how we store, index and search image and video collections several orders of magnitude larger than the Mouse Atlas but orders of magnitude smaller than our projected estimates for whole-brain scans. He’s also a long-time friend of Luc Vincent who directs the StreetView team.
I feel I could write a book about what I learned from Kevin and Mike and wouldn’t have scratched the surface of what they know that might be relevant to BAM. I have follow-up meetings scheduled and possibly a visit to Seattle to learn more about what’s going on in the Allen Institute. I have Kevin’s slides and he’s promised to upload his videos to YouTube. Mike, whom I just left an hour ago, said he’d send his slides and ask Christof for clearance to upload his talk to YT. I expect you know Kevin or met him when you were at NIH pitching BAM. (Note: After sending this message, I remembered Kevin telling me that he was the first student to enter Terry’s Computational Neurobiology Training Program at the Salk Institute.)
January 31, 2013
Here’s a note that I sent to Mehran Sahami who is the chair of the curriculum committee responsible for approving Stanford CS courses and any changes to their content:
Hi Mehran,
It’s that time of year when I start thinking about what I want to teach in CS379C. I intend to continue with the “scalable neuroscience” theme from last year and I’ve updated and re-purposed the lecture that I gave in Yoav’s class in November to highlight three technologies, Stephen Smith’s array tomography, Ed Boyden’s 3-D optical probes, and the connectomic technologies being developed by Winfried Denk and Sebastian Seung, as steps toward such scaling. I’m planning to invite Stephen, Ed, Sebastian and Kevin Briggman, formerly in Winfried’s lab and now starting his own lab at NIH, to participate in a manner similar to last Spring. Stephen and Kevin have already agreed and suggested some other scientists whom I know at Caltech and the Allen Institute who might be good to include.
This time around I will pay less attention to large-scale brain simulations — though I will spend some time discussing Markram’s HBP given its coverage in the press following the EU funding announcement, and more attention to the methods and instruments being developed for collecting data at brain scale. I want the students to learn to think quantitatively about neural recording and intevention technologies much as an engineer designing a cellular network might think about tradeoffs involving transmitter power, available frequency spectrum, the spatial distribution of cell towers and subscribers, etc. I want them to learn useful order-of-magnitude brain statistics and become adept at back-of-the-envelope calculations to determine for themselves what’s possible and what’s not.
I’d like to inspire them to become the instrument builders — which increasingly involve non-trivial algorithmic and computer-systems components — for the next generation of neuroscientists, serving as the modern day incarnations of the likes of Antonie van Leeuwenhoek and Ernest Rutherford. It would also be great to kindle their entrepreneurial zeal to found startups designed to solve specific problems while at the same time accelerating technologies that might allow us to vet more ambitious and detailed theories of brain function than currently possible. That’s a tall order and I’ll be happy if I can make a few steps in the right direction.
I wrote down a first approximation of an “advertisement” for the course in case you want to circulate it to the curriculum committee for their approval. I’m still working out the details of how students will be graded. As in years past, grades will be based on class participation, a midterm proposal produced in consultation with the instructor, and a final project, but this time I’m thinking of projects that entail a careful analysis of how, for example, a cell recording technology using a nanoscale optical-fiber network might scale to billions of neurons or how to manage the projected 50 billion terabytes of SEM data that would be produced by scanning a mouse brain, coupled with a business plan that might appeal to forward-thinking venture capital firm — there are actually some credible firms hunting around in this space. I’m collecting comments on the current draft and will produce a final version that I’ll ask my Stanford colleagues to circulate as the Winter quarter draws to a close:
In the version of CS379C offered in 2012, we focused on how to scale neuroscience to move beyond current technology for recording from small numbers of neurons to deal with much larger neural circuits. We also investigated novel investment strategies for funding science and financing startups that might accelerate the basic science and push the state of the art in related medical applications. In the intervening year, Henry Markram whose research at EPFL we studied headed a team of scientists who were awarded over a billion euros to fund the Human Brain Project (HBP) which Markram co-directs. The proposal was met with skepticism by many neuroscientists when it was submitted in 2012 and the announcement of the award in January of 2013 produced similar commentary, but it is clear that HBP aims at scalable neuroscience and now has the funding to pursue its goals.Your comments and suggestions on content or wording are welcome as always.Many scientists think a big-science approach to brain science reminiscent of the Large Hadron Collider (LHC) which took ten years and more than 3 billion euros just for the accelerator isn’t likely to be successful. Which isn’t to say they believe spending a lot of public money on brain science is a bad idea; most scientists do agree that a concerted effort to understand the brain will yield significant benefit to society. Indeed there is a consortium of scientists putting forward the Brain Activity Mapping (BAM) Project as a target of opportunity for US funding agencies including NIH, NSF and DARPA. BAM has a different agenda than HBP but no less grand challenges.
Imagine it is 2010 and the Human Genome Project (HGP) has yet to meet its goal of sequencing the human genome but the demand for sequencing is growing exponentially, new scientific and medical applications are surfacing every day, and there is money to be made. You are a partner at a venture capital firm interested in funding startups, or, if you prefer, a program officer at NIH interested in funding science to cure cancer. What projects would you back to maximize your impact? Now substitute BAM or HBP for HGP and Alzheimer’s or Parkinson’s disease for cancer. In CS379C this Spring, we will look at the enabling technologies for both HBP and BAM as a window on the future of scalable neuroscience and undertake the difficult task of predicting winners and losers in the key technologies of biomedical imaging, synthetic biology, recombinant DNA and nanoscale sensors and communication networks.
We will be looking at a broad swath of science and technology and students are not expected to understand all the contributing disciplines. This is exactly the situation faced by technologists working in areas contributing to brain science. Engineers in nanotechnology may not know a great deal about the biochemistry of neural signal transduction, but they’re willing to team with experts who do to build sensors for recording cell membrane potentials. Computer scientists may not know the details of the Hodgkin-Huxley neuron model, but they routinely collaborate with neuroscientists who do to build large-scale neural circuits. If you find this topic intriguing, you might check out this lecture presenting an overview of the approaches and component technologies we will be looking at in CS379C this Spring.
January 29, 2013
Biochemistry — which overlaps but does not subsume organic chemistry — is the study of chemical processes in living organisms and in particular the study of biochemical signaling and the flow of chemical energy in metabolism. Many if not most students nowadays get a basic introduction in high school and I’ve suggested in earlier posts that if you didn’t take chemistry in high school or want a quick refresher you might check out Eric Lander’s introductory lectures in the OpenCourseWare version of Introduction to Biology (MIT 7.012).
Realizing that you may not have the time or patience to follow this advice and in an effort to make these notes self contained, I would like you to understand the basic properties of ionic and covalent bonds. The most satisfying explanation that I’ve run across for the difference between ionic and covalent chemical bonds — along with a clear technical explanation for why the organization of the periodic table of elements is so useful — is in Chapters 11 and 12 of Michael Fayer’s book on quantum mechanics [55], but you can find a shorter explanation on chemistry.about.com — or check out this footnote67 — if you don’t have access to Fayer’s book.
January 27, 2013
I sent Ed Boyden an excerpt from the Briggman post summarizing what I learned from Kevin’s visit and the mentioning the monoclonal-antibodies nano-probe developed by the ORNL team led by Tuan Vo-Dinh. Here’s the preamble to my note minus the excerpt and followed by Ed’s reply:
TLD: Kevin Briggman formerly from Denk’s lab now at NIH running his own lab dropped by on Friday and we talked about what his new lab will be working on and about BAM as he was involved early on when George, Terry and others approached NIH. I’ve included my notes below if you’re interested in what we talked about. Relative to this message however, we talked about the limitations of connectomics and the importance of dense sampling individual cells in the process of inferring function. I got curious about nanoscale probes and did some research. As a meta comment, I found the field to be much larger than I expected, balkanized in all sorts of dysfunctional ways, divided into silos by academic-discipline boundaries and national-lab turf wars each with their favorite venues for publishing and conference attendance — in a word “baffling” — and difficult to survey given the wide variety of applications and nomenclature. Be that as it may, I now have a lot of new things to look at and try to understand.One thing that caught my eye concerned the clever combination of mononclonal antibodies and fiber optic probes for detecting specific molecules within cells. Do you know of promising work in this area? The references I did find seemed isolated — see Tuan Vo-Dinh reference below — and did not fill me with confidence that the ideas were well tested and understood, but the general approach does seem promising.
ESB: On the note about fiberoptic nanosensors — potentially, the two don’t have to be connected; we can modularly decouple if we can make fiberoptic brain monitoring arrays (like we talked about last time), and then also have nanosensors float around exhibiting fluorescence changes. This could be more modular, and even increase spatial resolution. However, in the short term, it’s great to have a platform for direct coupling: that maximizes signal to noise at a point, I would guess. A meta-theme here is that at the intersection of nanotechnology / hardware engineering + molecular engineering, and chemistry / molecular biology + imaging, is where the greatest progress might arise!
January 25, 2013
Kevin Briggman visited and gave a talk primarily on work he did while in Winfried Denk’s lab (Max Planck Institute, Heidelberg), but also looking forward to the new work he’ll be doing in his lab at NIH. He started out with a great analogy to his earlier work at Intel where they reverse engineered the chips of competitors using a variant of serial-section electron microscopy in which they repeatedly ablated the surface of the chip to reveal the various layers produced by lithography. Kevin pointed out that, in attempting to infer circuit function, the Intel engineers were aided by the fact that they pretty much knew what CMOS transistors and standard logic gates should look like even in the competition’s chips. Later he used to the analogy to point out the challenges in applying similar technologies to reverse engineer the function of starburst amacrine cells. His talk was full of interesting points, a few of which I’ve include here.
In discussing changes that occur during tissue preparation, Kevin mentioned that 25% of the volume of your brain consists of extracellular space primarily occupied by fluids, e.g., electrolytes. The extracellular matrix also includes connective tissues that support neurons68. This was a revelation to me as I had incorrectly concluded from looking at Sebastian Seung’s connectomic slides that cells were tightly packed with very little intervening space, even though this presented a problem for me in thinking about extracellular diffusion processes. When tissue is prepared for serial-section electron microscopy the chemicals used, e.g., formaldehyde, can alter the concentration gradients drawing water into the neurons, reducing the extracellular volume and making it more difficult to identify cell boundaries. Briggman is working with biochemists to find preparations that don’t have this property as well as exploring other alternatives for tagging synapses such as immunoflorescence.
Kevin’s NIH lab will be looking at mouse retina, zebrafish and eventually whole mouse brain. As in the work of Clay Reid, Denk’s lab used calcium imaging followed by SEM and were able to perform alignment by exploiting the preserved vascular structure. Kevin developed several effective protocols for segmenting cell bodies and tracking neural processes while working in Denk’s lab [8]. He is hoping that new tissue preparation methods and the increased resolution of the latest microscopes will help improve the automated techniques. He mentioned a new Zeiss electron microscope that achieves a substantial increase in throughput by splitting the beam into sixty-four sub regions and biasing the tissue sample to control light scattering. It sounds like Denk’s lab is getting at least one of these rather expensive machines. At one point, Chris Uhlik did a back-of-the-envelope calculation and estimated that a complete scan of a mouse brain would produce 50 billion terabytes of data at one bit per voxel69. You can find the PDF for Kevin’s slides here and he promised to upload his videos to YouTube. He also agreed to participate in CS379C in the Spring.
Briggman agreed that Tony Zador’s proposal [172] is perhaps the most realistically near-term scalable BAM [6] technology, but he was skeptical that simply having the adjacency matrix for the connectome would turn out to be very useful. He pointed out that the locations where one axon connects via a synapse to a dendrite and whether that connection is excitatory or inhibitory is crucial for understanding many if not most neural circuits. We also talked about the prospects for using a variant of Stephen Smith’s array tomography [109] coupled with some tricks from immunohistochemistry70 so that all four of the major neurotransmitters could imaged using a single preparation thus avoiding some of the alignment problems that occur with multiple preparations. On a mostly unrelated note, I found out that the basic principle guiding the design of the confocal microscope was patented in 1957 by my academic great grandfather, Marvin Minsky71.
The Google Brain folks got a DistBelief version of Alex Krizhevsky’s [91] convolutional network for the ImageNet task trained and outperforming their previous top-performing network [94]. Krizhevsky’s highly-optimized CUDA implementation is currently faster than DistBelief. Yann Lecun explained the performance of the Krizhevsky et al [91] network in terms of the following features:
a super-fast implementation of convolutional nets on dual GPUs with a network architecture designed to just fill up the memory of the two GPUs, exploiting parallelism by training on mini-batches of a couple hundred of samples so the network takes about a week to train,
rectified-linear units — if x < 0 return 0 otherwise return x — combined with max pooling for the non-linearities,
contrast normalization, but only across features — not across space which apparently doesn’t help,
lots of jittering, mirroring, and color perturbation of the original images generated on the fly to increase the size of the training set,
a few hacks to initialize the weights, prevent the weights from blowing up for large values of the learning rate, and
a “drop-out” trick for the last two layers in which half the units — picked at random — are set to zero during training, so as to make each unit more independent from the others.
The analysis of the top two performers in the Imagenet Challenge — Krizhevsky et al’s SuperVision from Geoff Hinton’s lab at the University of Toronto and Simonyan et al’s VGG from Andrew Zisserman’s lab at Oxford University — including a comparison of the PASCAL and LSVRC (Large Scale Visual Recognition Challenge) is worth a close look if you’re working on object detection or interested in the evolution of the image datasets that are driving the field.
Ran across an interesting nanoscale sensor-probe technology in a review article by Vo-Dinh and Zhang [159]. The most intriguing work was carried out by a team of scientists at Oak Ridge National Labs where I found a press release describing the probe which is referred to as a nano-needle in the following excerpt:
The nano-needle is really a 50-nm-diameter silver-coated optical fiber that carries a helium-cadmium laser beam. Attached to the optical fiber tip are monoclonal antibodies that recognize and bind to BPT72. The laser light, which has a wavelength of 325 nm, excites the antibody-BPT complex at the fiber tip, causing the complex to fluoresce. The newly generated light travels up the fiber into an optical detector. The layer of silver is deposited on the fiber wall to prevent the laser excitation light and the fluorescence emitted by the antibody-BPT complex from escaping through the fiber.There were also some possibly related papers on Vo-Dinh’s web site at Duke University, but there were no PDF preprints aside from conference posters — see here and here — and I didn’t have time to pursue using my Stanford or Brown University library privileges.This nanosensor of high selectivity and sensitivity was developed by a research group led by Tuan Vo-Dinh and his coworkers Guy Griffin and Brian Cullum. Using these nanosensors, it is possible to probe individual chemical species and molecular signaling processes in specific locations within a cell. We have shown that insertion of a nano-biosensor into a mammalian somatic cell not only appears to have no effect on the cell membrane, but also does not effect the cell’s normal function. (source)
Here are three possible solutions for the GPU “math library” that Dean Gaudet and I discussed in our last conversation:
ArrayFire C++ library developed AccelerEyes and providing a similar functionality to their Jacket product for GPU accelerated Matlab functions.
Theano — License: BSD — contains some NumPy code — see here for details.
PyCUDA and PyOpenCL — Licence: MIT/X Consortium — excludes some components that fall under terms of Thrust — see here for details.
I talked with Dumitru Erhan and he said that he is going to port Theano and will get in touch with Dean about EASYMAC access. I also talked to John Melonakos, CEO of AccelerEyes and he gave me two ArrayFire licences to experiment with which I advertised to DistBelief if anyone was interested in taking for a spin.
Michael Frumkin told me about work he has been following at the Gladstone Institutes on Molecular Mechanisms of Plasticity and Neurodegeneration led by Dr. Steven Finkbeiner.
January 21, 2013
While at the Kavli Futures meeting, I talked with David Heckerman, who is an expert in probabilistic graphical models and their applications in bioinformatics. David had some interesting ideas about using ultrasound as a means of neural readout. I would have been more skeptical except for what one of my former students has been telling me about new ultrasound technologies for probing deep tissue. Prompted by David’s suggestion I did a literature search and discovered a wealth of technological innovation in applying ultrasound to sensing and imaging.
Contrast enhanced ultrasound (CEUS) combined with ultrasound based contrast agents has been used to enhance traditional medical sonography. Functional Transcranial Doppler Spectroscopy (fTDS) is a neuroimaging tool for measuring cerebral blood flow velocity changes due to neural activation during cognitive tasks. There are also a host of new photoacoustic spectroscopy technologies that combine ultrasonic and electromagnetic radiation to enhance imaging73.
My former student, Chi-Yin Lee, now works at Siemens Medical Systems in the Ultrasound Group. CY is currently working on High-intensity Focused Ultrasound (HIFU) technology to treat cancer. Siemen’s competitors including Philips and GE already have products in this space and it is highly competitive. The Foundation for Focused Ultrasound Research (FFUS) is an American non-profit 501(c) charity designed to promote both education and research within the area of medical applications of focused ultrasound. Current methods use fMRI to locate the tumors and the ultrasound the ablate them, but the holy grail is to use ultrasound for both ablation and localization. CY suggested that we might want to look at so-called targeted microbubbles in engineering various sensing neural-state technologies74.
At Caltech, I met with Akram Sadek for an hour prior to the Kavli meeting to learn what he’s been up to. After a two-year hiatus during which he had to do a rotation in a biology lab he’s back in Axel Scherer’s Nanofab Lab and working on some new ideas for improving the S/N ratio for his cochlea-inspired NEMS multiplexer. Akram thinks that the solution may require using some alternative to CMOS for his design, but he’s also exploring some other workarounds. We talked about reading membrane potentials using quantum dots and carbon nanotubes, e.g., Kawano et al [81] and Xiaojie et al [52] for examples of nanotube-based neural-sensor technologies and Fabbro et al [54] for nanotube-based neural-stimulation technologies, and using pairs of QDs and FRET to build a sensor interface to a micron-scale optic fiber.
January 19, 2013
Fritz Sommer, Larry Swanson and I got to talking about challenges of mapping brains and registering landmarks on a sample brain. When Bruno Olshausen, Fritz and the Redwood Center were thinking about the neuroscience data project, we offered to write a letter of support to NSF and provisionally agreed to help out with managing the data, but that never happened in large part due to there not being much data but also to lack of consensus among the participants regarding what constitutes appropriate provenance. At the time we talked about extending Google Body which had a rough map of the nervous system and the capacity to render with greater precision if there was good data. Yesterday I mentioned to Fritz and Larry that Google Body is now an open source project and Zygote Media Group, which provided the imagery for Google Body, has used this open source code to build Zygote Body which runs in several popular browsers. If BAM started collecting lots of 3-D images of brains and other neural tissues, we might be interested in helping out. Google Body was done on 20% time and I would be surprised if we couldn’t find some talented engineers to rise to the challenge of developing interactive tools for exploring the brain. The related problem of taking a piece of brain tissue annotated with a set of landmarks and registering it with one or more “standard” samples is likewise liable to attract some engineers who work on Google Earth, StreetView, Maps or anyone of a half dozen other Google products that involve computational photography and cartography.
On Thursday, I had a lot to say about how to manage huge digital repositories, from my team’s experience in sharding, compressing, curating and indexing billions of videos for YouTube and some personal experience in a previous lifetime with the repository at NASA Goddard where much of the data from the Earth Observing System (EOS) resides and is made available to scientists around the world. EOS is particularly interesting for the heterogeneity of the space craft and their sensors and for the challenges involved in co-registering the resulting data for all sorts of scientific studies not the least important being the role this data has played in climate science. When the repository was first proposed, the engineers had the foresight to realize that there was no simple solution to the problem of curating and cataloguing the data as no one could anticipate how the data would be used. They also had the problem that Google has and BAM probably will have namely that once the fire hose is turned on there will be few options other than cataloguing the data as it is transmitted from the satellites or uploaded in the case of video. Twenty years ago when the Goddard facility was built they designed an elaborate storage hierarchy with the last resort for infrequently accessed data being a massive tape storage facility where robot librarians would find and load a tape containing a specific dataset if a scientist required it. As you might guess, this alternative was quickly rendered obsolete by faster, higher-capacity storage technologies. Most of the original satellites hardly qualify as fire hoses today, but each new generation has posed new challenges for the Goddard computer scientists.
Finally, in conversations with Clay Reid, Fritz and John Doyle, I described how venture capital firms in silicon valley have accelerated genomics by funding startups in both hardware and industrial-scale systems for sequencing and applications and value-added services. There is no doubt in my mind that this infusion of cash from multiple VC firms has considerably sped up the march to the $1,000 genome. After an aborted attempt in the late nineties and early aughts to find commercial applications for research in neuroscience, VC firms are again looking for opportunities in the brain sciences, this time led by partners with deeper expertise in bioinformatics, medicine, neuroscience and computational scaling. Perhaps this time around they won’t get fooled by the lure of enthusiasts who think they know enough to build a brain. I pointed out that, while we don’t know how to build a nanoscale communications network to read off neural state at scale, we do have lots of interesting ideas and promising component technologies. While well short of BAM’s grand challenge, there are some interesting intermediate challenges in personalized medicine, biocompatible sensor networks and even entertainment that may have near-term application and that could serve to bootstrap key enabling technologies for BAM’s longer-term goals. I suggested that we make it a central focus of BAM to work with entrepreneurs and investors to identify such opportunities, seek out and encourage talented innovators and assist with matching inventors and investors.
January 15, 2013
In anticipation of the opportunity of talking with him at Caltech on Thursday, I read Clay Reid’s paper analyzing the relationship between structure and function in networks consisting of thousands of neurons by using a combination of two-photon calcium imaging to infer preferred stimulus orientation of a group of neurons in the mouse primary visual cortex and large-scale electron microscopy of serial thin sections to infer local connectivity [20]. The paper is perhaps more interesting for the discussion of methods than for the conclusions drawn. Reid recently moved from Harvard to take a position at the Allen Institute for Brain Science.
Array tomography as a means of 3-D proteomic imaging is another potentially scalable neural sensing technology [109]. Here is a description of array tomography from Stephen Smith’s Lab:
Array tomography (AT) is a new high-throughput proteometric imaging method offering unprecedented capabilities for high-resolution imaging of tissue molecular architectures. AT is based on (1) automated ultrathin physical sectioning of resin-embedded tissue specimens, (2) construction of planar arrays of serial ultrathin sections on optical coverslips, (3) staining and imaging of these arrays, and (4) computational reconstruction into three dimensions, followed by (5) volumetric image analysis. Array can be imaged by immunofluorescence or by scanning electron microscopy, or both. Because these arrays are very effectively stabilized by the glass substrate, they can withstand many repeated cycles of staining, imaging and elution. This permits the imaging of large numbers (i.e., 20 or more) of antibodies in addition to ultrastructure on each individual section. (source)
January 13, 2013
In the last entry, we considered possible molecular machinery for positioning optical fibers and ended with the suggestion that we might finesse the problem entirely by creating a scaffold with the optical-fiber network already in place and grow the neural network around the existing communications network. This move would simplify the problem of how to attach the sensor-encoder-light-transmission package in situ by allowing us to attach the packages prior to positioning. I’ve read that aligning fibers to wave guides in the assembly of fiber optic networks is difficult and it would seem that it will be no less challenging at the nanoscale and in the case of self-assembly in particular.
In this entry, we briefly mention some ideas relating to the sensing, encoding, multiplexing and light-emitting packages that might be attached to each fiber. Even if we are able to insert fiber optic cables deep into neural tissue, it may be impractical to use one cable per sensor. In Using piezoelectronics to wire thousands of neural nanosensors into a single optical output (HTML), Akram S. Sadek, a graduate student at California Institute of Technology’s Computation & Neural Systems program describes a nanoelectromechanical-systems (NEMS) device that he claims can multiplex thousands of neural nanosensors using a nanoscale analogue of the cochlea — or at least our current theories of how the cochlea works75. Ed Boyden was familiar with the paper [133] and had talked with one of the coauthors, Sotiris Masmanidis, who reported at the time that the signals were very small requiring them to average over time to get reliable readings. I sent email to Sadek asking him for an update and will try to follow up when I’m at Caltech on Thursday.
I ran across Nanoscale electrical and optical sensing of vertebrate and invertebrate cultured neural networks in vitro (HTML) by Ronald Adam Seger who is now in Professor Nader Pourmand’s Biosensor & Biotechnology Group at the University of California, Santa Cruz. The document was only available for a price through a dissertation reseller but the abstract and free 24 page preview wasn’t enticing enough for me to spring for the full document. In reading the preface of Seger’s thesis, it occurred to me that it might make sense to design our sensor-encoder-light-switching packages so that we transmit signals by modulating incoming light and reflecting it back along the same incoming path. I’m sure this would have occurred immediately to any communications engineer and it may make sense in our case to have the photon source external to the neural tissue to reduce internal power requirements.
I am also trying to get up to speed on the applications of quantum dots as electro-optical modulators and switches. In reading an IEEE Spectrum article on quantum dots as single-photon emitters in communication networks, it became clear to me that I have no idea how practical current techniques are for our applications. If anyone knows of a good survey article, I’d appreciate a reference; Wikipedia has helped but I could benefit from a more in-depth tutorial presentation.
On a mostly unrelated topic, I’ve been collecting animations to illustrate various points in my introductory lectures for this coming Spring’s instantiation of CS379C; here are best animations that I was able to find for two topics that I frequently have to cover and that are particularly well communicated using a self-paced animation:
Two animations illustrating synaptic transmission: one at Harvard’s Department of Molecular and Cellular Biology (SWF) and one at McGraw Hill’s Online Learning Center (HTML).
Two animations illustrating action potential propagation: one at Harvard’s Department of Molecular and Cellular Biology (SWF) and one at W. H. Freeman’s Online Learning Center (SWF).
I like to contrast chemical junctions with electrical junctions implemented by gap junctions which allows me to introduce dendro-dendritic neural circuits (SWF). It is worth pointing out that gap junctions serve different purposes and occur in many different cell types in addition to neurons and glia.
I was also looking for a relatively high-level introduction to or review of the central dogma of molecular biology. I didn’t spend much timing looking and I wasn’t entirely happy with what I came up with but I’m assuming that most students are familiar with the basic steps — DNA to RNA to protein — and so will either skip this or use it for a quick review. You can find my picks here and here.
January 11, 2013
Spent an evening curled up with Google Search experimenting with queries like “inserting optical fibers in neural tissue”. Among other interesting documents, I found an OpenOptogenetics Wiki and a bunch of papers with titles like “Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits” and “Fiber-optic implantation for chronic optogenetic stimulation of brain tissue” which, while interesting for the information relating to biocompatability, are primarily focused on optogenetic stimulation and have a shorter horizon — around two weeks — for “long-term” than I was thinking of. If we are to seriously consider Chris’ proposal we’ll need to know a lot more about the biocampatability and biodegradability of naked optical fiber, the tensile strength for initial positioning and subsequent stress due to changes in the neural tissue, as well as available nanoscale components — linear and rotary motors — for positioning optical fibers. Frustrated with what I could find on the web, I went back and re-read — this time more carefully — the relevant chapters of [76].
Chapter 6: Machines and Mechanisms in Jones [76] provides a useful review of what is known about molecular motors and our attempts to utilize them or build synthetic versions. He describes how molecules of myosin and actin in our muscles work [138] together with Brownian motion and the hydrolysis of ATP to change the shape and stickiness of the molecules to produce motion76. Sal Khan provides a nice explanation of the relevant chemical and kinematic processes in one of the biology lessons offered by the Khan Academy. The same principles — conformational change and switchable stickiness coupled to the catalyzed hydrolysis of ATP, with the shape changes driven by Brownian motion — apply to a wide range of linear motors found in nature. In particular, we already looked at the cellular transport mechanisms involving microtubules and the transfer molecules kinesin and dynein.
In addition, there are developmental transport mechanisms responsible for the final positioning of cells in which, starting from sites of neurogenesis, migrating neurons climb scaffolding consisting of the processes of radial glial cells to find their final locations in the developing brain. In the Spring 2012 instantiation of my Stanford class, we looked at how growing axonal processes follow molecular gradients during the development of retinotopic maps in the visual cortex [108]. In terms of synthetic motors, Hiroyuki Noji and Masasuke Yoshida [171, 72] were able to “isolate rotary motors from bacteria, anchor them to a surface and then attach protein threads, 4 microns in length, to their rotors.” When they added ATP to the solution, they could see the threads rotating by observing them with an ordinary light microscope (source: page 150 [76]).
Despite some progress in developing synthetic motors, we are still just starting to understand the basic principles and quite some way from figuring out how to create fleets of synthetic motors, marshal them via self-assembly to distribute and attach themselves to optical fibers and then snake these fibers through a dense tangle of axons, dendrites and cell bodies into positions within the brain, there to be attached to sensors for sensing and converting information to light pulses. As an intermediate step, it would be interesting to consider how we might got about creating a scaffold, or organic matrix in which to grow neural progenitor cells and create a small neural network with the optical fiber network already in place? Jones also includes an interesting discussion of biosensors with a detailed explanation of piezoelectric devices in which changes in voltage produce changes in shape — specifically, the conformation or proteins — and, conversely, changes in shape produce changes in voltage.
January 10, 2013
I’ve been following progress on the therapeutic applications of TMS (transcranial magnetic stimulation) which is interesting in part because it is a relatively non-invasive method of neural stimulation with few side effects. Yesterday Brainsway Limited a medical device maker based in Israel announced that it has obtained US Food and Drug Administration (FDA) clearance for use of its Deep TMS device for the treatment of depression in patients who fail to respond to therapeutics during a depression episode. The company adds that the FDA approval for this indication is generally broader than the definition given by the company’s TMS device rival. Their technology was described in this recent article in IEEE Spectrum.
In thinking about the microbial opsins used in optogenetics to investigate the function of neural systems, e.g., channelrhodopsins, I got curious about other sources of bioluminescence and stumbled across Edith Widder’s TED talk on bioluminescence in sea life. Widder claims that, if you drag a net from 3,000 feet to the surface, most animals — in many cases 80–90% of the animals — make light. They do it to identify food, attract prey, find mates and confuse or distract animals that prey on them by ejecting bioluminecent material, in some cases not just Luciferin but whole cells with nuclei and intact membranes, at considerable metabolic cost.
January 9, 2013
I had asked Chris Uhlik for his quick back-of-the-envelope evaluation of the feasibility of scalable wireless neural-state readout, and here is his response:
Wireless Radio: 1e10 neurons × 1 bit per msec, 50% overhead for packetization, addressing, error correction, etc. is 1.5e13 bps or 15000 Gbps. Clearly it cannot be transmitted through a single channel with anything like a few GHz of bandwidth at any SNR; it must be transmitted from many transmitters and received by many receivers. Transmitters that are close together cannot act in a completely decoupled manner. Conventional practice keeps transmitters separated by at least 1/4 wavelength and better 1/2, but let’s go with 1/4 wavelength and 1 GHz. Higher frequencies are absorbed by the salty water brain. I’m going to use 1 GHz of bandwidth at 1 GHz for round numbers 500MHz to 1500MHz and limit the spacing of transmitters to 1/4 the wavelength of 500MHz. Oops, the wavelength of 500 MHz is 6 inches. You can’t fit two of them inside your skull at once. This isn’t a radio frequency transmission problem. Since it is near field, you probably can use various effects that I don’t understand to pack them tighter, but still we are talking maybe 10 × 10 × 10 rather than billions of them. Still 10 × 10 × 10 gets use down to about 15 Gbps per transmitter. Fitting 1000 receivers around the surface of a 6 inch sphere puts them 1 mm apart, so right away we have a problem that we cannot fit anywhere near 1000 receivers on your scalp while keeping their antennas at all independent.Near-field communication which is used in RFID tags and smartphones has some of the same baud rate and frequency spectrum issues that Chris mentions but is another possibility worth looking into as we consider nanoscale network devices. A near-field magnetic induction communication system is “a short-range wireless physical layer that communicates by coupling a tight, low-power, non-propagating magnetic field between devices. The concept is for a transmitter coil in one device to modulate a magnetic field which is measured by means of a receiver coil in another device” (source). Near-field communication (NFC) refers to a set of standards for smartphones and similar devices and used to implement such services as Google Wallet.Physical Transport: If you can tolerate a long readout delay, perhaps the encoders can write the data onto RNA strands and pack them into bacteria-like cells which are dumped into the bloodstream and filtered out elsewhere. This is a physical transport of the bits approach which is sort of like using FedEx to send disk drives across the country. A box of 50 × 2TB disks delivered in 24 hours is 10 Gbps.
Fiber Optic Cable: Let’s look at an energy balance assuming that bits are sent using unmodulated photons (1 photon per bit, clocking magically reconstructed with a few extra bits). Let’s use photons that are 5 microns long, about the length of a typical bacterium. 15,000 Gbps × h nu = 600 nanowatts. If you can get the photons out of the brain without being absorbed, the power requirements don’t look bad. Communication is possible with just a few hundred photons per bit. If you could riddle the brain with 8 micron core mono-mode optical fibers, you could get the bits to the surface. Suppose you could terminate 1 of these fibers per cubic mm of brain tissue and keeping with the 6 inch spherical brain model, we have 1.8M fibers utilizing ~0.2% of the scalp area. The transmitter machine needs to collect data from 5400 local neurons up to 1/2mm away and send 5.4M bps up the fiber.
In far-field transmission of the sort used in most WIFI technologies, all of the transmission energy is designed to radiate into free space and the power density attenuates at a rate proportional to the inverse of the range to the second power. By contrast, near-field transmission works by using magnetic induction between two loop antennas located within each other’s near field, effectively forming an air-core transformer; the power density of near-field transmissions is extremely restrictive and attenuates at a rate proportional to the inverse of the range to the sixth power resulting in a working distance of about 4–6 centimetres.
Separate from his response to my back-of-the-envelope query, Chris and I met and briefly chatted about an idea he had for creating a optical fiber network in the brain. It sounded interesting and I asked him if I could forward it to Ed Boyden which he encouraged and so I did. Here’s the note that I sent to Ed, including Chris’s idea and Ed’s comments in line:
Discussion concerning the challenge in the Alivisatos et al [6] and Terry Sejnowski’s white paper have engendered a lot of discussion. Some of our engineers have done the back-of-the-envelope calculations and can’t see any viable technologies on the immediate horizon for the wireless readouts that George envisions. Various schemes for encoding state and flushing appropriately encapsulated cell products into the blood and eventually urine have been proposed but that doesn’t seem to capture the imagination like a real-time readout technology.ESB: Agreed. I’ve run similar calculations for bandwidth (mouse: 50 million neurons, 20 kHz, etc.) and it’s going to be hard.
Chris Uhlik who has great track record for novel solutions involving acquisition of geographic data by StreetView vehicles, aircraft, satellite and autonomous robots for indoor applications, came up with an interesting idea that caught my fancy and I wondered if you knew of anything related, directly or otherwise. The ideas is to wire a brain with micron scale fiber optic cables using some sort of — and here’s the really hand-wavy, wishful thinking part — diffusion, or gradient-following distribution process. Kind of wild but there’s something about the proposal that keeps nagging me. I was hoping you had some tips about where to look for leads or perhaps a different way of thinking about the more general idea. Here’s Chris’s original idea which I’m forwarding to you with his permission:
Chris: I think outfitting a brain with optical fibers may in fact be quite reasonable. Depending on diffusion or EM transport through ion-filled water seems like a losing proposition. But guided transport of photons though optical fiber is practically perfect. And, the functional core of optical fibers can be quite small in diameter. Neurons are already hooked together by axons and dendrites which act like a sort of EM wave guide. I would like to see work on technologies to insert/guide optical fibers through a healthy brain while minimally disrupting existing connections. Sort of like horizontal oil drilling, it might be possible to twist the fiber and have some molecular machine at the fiber tip guide the tip over/under/around existing structures, pushing them gently out of the way. The machine at the tip has the benefit of energy and communication with the outside world. Inserting a few xxx thousand of these and guiding them to desired places might enable sampling a few points in every cortical column. The energy demands are completely within bounds because optical fiber is so much more efficient a transport conduit than an axon.ESB: Sure! We’ve already started building massively parallel 5-10 micron cross section 3-D optical fiber devices, using optics microfabrication strategies, and we are aiming to tile the brain as densely as we can with them [176]. We’ve implanted about a dozen so far into living mouse brain. It’s a great idea! Want to forward the above paper to your colleagues, and we could brainstorm further?
January 7, 2013
In reading an article [158] that surveyed recent applications of nanotechnology to medicine, I learned about the CombiMatrix lab-on-a-chip which was also featured in Biochips As Pathways To Drug Discovery by Andrew Carmen and Gary Hardiman.
CombiMatrix DNAarray tests are lab-on-a-chip devices for identifying and investigating genes, gene mutations, and proteins. These miniaturized, microfabricated, semiconductor biochip-based systems are capable of performing standard or customizable multiplexed assays involving DNA, RNA, proteins, peptides, or small molecules, so they have many potential applications. The first CombiMatrix Diagnostics product was introduced in 2005 (source).On a related note, the microfluidics-based technologies used by the V-Chip are quite interesting and worth learning about to round out your bag of nanoscale-enabled personalized-medicine technologies that might yet turn out to be relevant to brain research [141].
In our conference call this morning, Krishna Yeshwant told me that he’s been talking with some researchers about exosomes which are 30 to 90 nm vesicles secreted by a wide range of mammalian cell types. I generally think about vesicles as holding bins for neurotrasmitters in axon termini that participate in communication between neurons by merging with the cell membrane and dumping their payload of neurotransmitters into the synaptic cleft. More generally “a vesicle is a small bubble within a cell, and thus a type of organelle. Enclosed by lipid bilayer, vesicles can form naturally, for example, during endocytosis (protein absorption). Alternatively, they may be prepared artificially, when they are called liposomes” (source). Turns out that exosomes can be used for all sorts of purposes:
Scientists are actively researching the role that exosomes may play in cell-to-cell signaling, hypothesizing that because exosomes can merge with and release their contents into cells that are distant from their cell of origin, they may influence processes in the recipient cell. For example, RNA that is shuttled from one cell to another, known as “exosomal shuttle RNA,” could potentially affect protein production in the recipient cell. By transferring molecules from one cell to another, exosomes from certain cells of the immune system, such as dendritic cells and B cells, may play a functional role in mediating adaptive immune responses to pathogens and tumors. Conversely, exosome production and content may be influenced by molecular signals received by the cell of origin. (source)Krisnha mentioned that he consults with Xandra Breakefield who is a Professor of Neurology at Harvard Medical School and suggested we might want to look at her work in thinking about alternative methods for intercellular communication. For example, excerpting from the abstract of Shao et al [137]:
Glioblastomas shed large quantities of small, membrane-bound microvesicles into the circulation. Although these hold promise as potential biomarkers of therapeutic response, their identification and quantification remain challenging. Here, we describe a highly sensitive and rapid analytical technique for profiling circulating microvesicles directly from blood samples of patients with glioblastoma. Microvesicles, introduced onto a dedicated microfluidic chip, are labeled with target-specific magnetic nanoparticles and detected by a miniaturized nuclear magnetic resonance system. Compared with current methods, this integrated system has a much higher detection sensitivity and can differentiate glioblastoma multiforme (GBM) microvesicles from nontumor host cell-derived microvesicles. We also show that circulating GBM microvesicles can be used to analyze primary tumor mutations and as a predictive metric of treatment-induced changes. (source)The review paper by Lai and Breakefield [92] entitled “Role of Exosomes/Microvesicles in the Nervous System and Use in Emerging Therapies” provides a better starting point for learning about exosomes and microvesicles, but I suspect you’ll need a good medical dictionary or a browser open to Wikipedia to understand many of the technical terms. The review paper, in addition to surveying the wide range of extra-cellular membrane vesicles (EMVs) and their related mechanisms in neurons — see Figure 1, describes current EMV-based technologies — see Figure 2 — and their targets including cancer immunotherapy and the use of EMV-technologies as the basis for in vivo targeted delivery of therapeutic drugs.
January 5, 2013
A couple of references in need of an anchor page: I think it was David Konerding who posted Neil deGrasse Tyson’s answer to the question “What is the most astounding fact you can share with us about the Universe?” on Google+. In any case, Neil’s answer deserves some contemplation and the fact — technically only a hypothesis — that he shares is certainly worthy of awe as is the realization that human beings were able to infer such a fact while still tethered to their home planet.
The title of this article says it all, “Promising Compound Restores Memory Loss and Reverses Symptoms of Alzheimers”, except that the study reports early results and only in mice. The mice receiving injections of the experimental compound (TFP5) “experienced no weight loss, neurological stress (anxiety) or signs of toxicity.” The comment, “next step is to find out if this molecule can have the same effects in people, and if not, to find out which molecule will”, indicates that, while it is possible TFP5 narrowly targets the specific Alzheimer’s phenotype found in mice, knowledge of the positive result in mice coupled with the molecular characteristics of TFP5 may give us enough of a hint to find a compound that works on the Alzheimer’s phenotype that afflicts humans.
Note to Krishna Yeshwant in anticipation of our phone conversation on Monday:
Looks like we’re scheduled to talk Monday afternoon. I maintain a research log for each quarter to keep my team up to date on research projects that we’re involved with in some capacity. The first two entries for 2013 Q1 are available here and are relevant to the Church proposal and Sejnowski’s white paper which we’ve been discussing.
January 3, 2013
I’ve spent a good portion of the last few weeks trying to better understand the technological problems that need to be solved to address the challenges put forward in the Alivisatos et al [6] paper in Neuron. As mentioned in an earlier post, the Zador et al [172] proposal addresses one instance of the Alivisatos et al challenges that I believe can be solved in two to five years. The next level of difficulty involves recording more than just connectivity, e.g., protein products, transcription rates, concentrations of calcium ions, spike-timing data. In the near term, I would bet that the sort of data we’ll soon be able to get from Boyden’s 3-D sensors will drive the search for a better understanding of individual neurons and small ensembles, and may even lead to significant medical discoveries. The connectomic data that a working version of Zador et al will provide should nicely complement Boyden’s 3-D data.
To move beyond implantable arrays, we need to figure out how to encode information concerning protein products and action potentials along with relevant timing information. The hope is that this can be accomplished by adapting existing biological components; the challenge is doing so within a metabolic budget that doesn’t so impoverish the cell that it ceases to function normally. Once we can encode information relating proteins products, action potentials and the like, and store it in biocompatible structures within the brain, the next step will be transferring that information outside the brain for analysis. This is the hardest problem posed in the Alivisatos et al paper, and I’m reasonably confident that relying on the near-term development of nanoscale communication is just wishful thinking — specifically, I believe a solution will require self assembly of a sort well beyond the current state of the art.
Even in terms of assembling the individual biocompatible components such as oscillators, packet routers and resonant transformers for near-field wireless power transmission we have a long way to go. The fact is that practical nanoscale device development falls far short of the hype that is reported in the PR releases from universities and industrial research labs that feed the tech tabloids’ — Engadget, Slashdot — insatiable appetite for sensational high-tech stories. In particular, assembling useful devices from existing components has had few notable successes. Even the reported successes, like single-atom transistors, are far from sort of components that an engineer can build upon to construct reliable memories and logic gates. It seems a good bet that building a nanoscale communications network to transmit neural-state data out of the brain is at least ten years from reality and it might be twice that estimate.
Babak Parviz suggested that the best way to accelerate progress is to focus attention on a particular problem to solve, much as attention is currently focused on high-throughput, low-cost gene sequencing. The ideal such problem would have a substantial economic payoff for the developer and would be somewhat less challenging than engineering brain-scale wireless communication networks. Perhaps as suggested in [27] the market for implantable biosensors [151] will serve to bootstrap the component technologies we need, but I’m skeptical since most of the current applications, e.g., monitoring intracranial pressure or treating Parkinson’s disease using neurostimulation — see Ventola [157, 158] for a recent review of medical applications of nanotechnology including toxicity issues, can manage without a distributed communications solution using components that are quite large compared with the requirements for nanoscale networks.
In the meantime, I think we have to focus on alternative and likely more cumbersome methods for scalable read-out. Babak liked the idea that I proposed in my Stanford lecture for encapsulating coded data in capsid-like structures and then flushing them into the circulatory system to be recovered by a filtration system like those used in kidney dialysis machines. But he suggested the even simpler expedient of creating information capsules that would directly marshal the normal filtration capabilities of the kidneys77 to flush the data-laden cargo into the urinary tract where it would be easier to process. In the case of lab animals like mice, a catheter could be used to collect the urine, or, simpler yet, use a nonabsorbent bedding material in the animal’s cage with a removable screened collection tray.
December 29, 2012
When Feynman discussed assembling nanoscale machines, he would often speak of first building a set of 1/4 scale tools, using them to build 1/16 scale tools, and so on, ultimately constructing millions of entire nanoscale factories. This leads some to think that nanoscale assembly will look like macroscale assembly — tiny machine tools made out of rigid parts, constructing nanoscale products out of materials that behave like the materials we encounter in everyday life. If we were to proceed with this intuition, we would very likely end up being disappointed. Nanoscale fabrication and assembly present new engineering challenges precisely because different physical laws dominate at different scales, but it also offers powerful new opportunities for combinatorial scaling.
Objects at the nanoscale, organic molecules in the case of biological systems, tend to be “flexible”, “sticky” and perpetually “agitated.” “Flexibility” refers to the fact that proteins and other large molecules that comprise biological systems generally have multiple shapes or “conformations”. Even once proteins are folded into a particular conformation, the geometric arrangement of their constituent atoms changes in accord with the attractive or repulsive forces acting between parts of the protein, e.g., Van der Waals force, and interactions with other molecules in their vicinity, e.g., due to the forces involved in making and breaking covalent bonds.
“Stickiness” refers to the fact that these molecules routinely exchange electrons allowing new molecules to be formed from existing molecules by way of chemical reactions catalyzed by enzymes. These molecules have locations — the “sticky” sites — corresponding to molecular bonds where electrons can shift their affinity to create new bonds with other nearby molecules — and hence “stick” together. Finally, “agitated” refers to the fact that the molecules are constantly in motion due to changes in conformation, interaction with other macromolecules, and being struck by smaller fast moving atoms and molecules. The attendant forces cause individual particles to undergo a random walk, with the behavior of the ensemble as a whole referred to as brownian motion.
In nanoscale engineering, these properties of nanoscale objects can channeled to create products by self-assembly. The study of soap films provides a relatively simple introduction to the natural processes involved in self-assembly, and there are a number of popular books in the library that detail these same processes at work in biological systems [50, 76]. Physicists like to joke that you don’t study quantum mechanics to understand it — since that is clearly impossible, only to apply it — and, of course, that implies a book on quantum theory that doesn’t include a lot of worked-out examples and derivations is of little value. Quantum mechanics is definitely a prerequisite for many nanoscale engineering applications, but it is also necessary to acquire intuitions that enable us to imagine how molecules interact both in pairs and in larger ensembles at these unfamiliar scales. Fortunately, biology provides us with a diverse collection of molecular machines we can study to develop those intuitions.
December 27, 2012
Recently we met with Dr. Uzma Samadani who is a neurosurgeon at the NYU Langone Medical Center. Dr. Samadani came to my attention recently when I learned that she was working on diagnostics for brain injury in collaboration with David Heeger who is the PI for the Computational Neuroimaging Lab at NYU. I knew David in graduate school and he and I used to go windsurfing together when he was on the faculty at Stanford. What’s novel about their approach is that it is non-invasive, relies on off-the-shelf technologies — basically eye-tracking, and in several surprising cases it performs better than expert diagnosticians and detects abnormalities, e.g., concussions, that more expensive and complicated procedures such as MRI and PET scans miss. Moreover, the necessary hardware can be assembled using a laptop and camera or, better yet, using HUD technology based on Google Glass.
How many other such diagnostic windfalls might be enabled by smart phones using their existing sensors or relative simple USB-tethered extensions? Eric Topol in The Creative Destruction of Medicine listed some of the related smart-phone applications like those used to detect glaucoma or infer a patient’s eyeglass prescription. Some of the more ambitious apps like the Zeo Sleep Manager — which requires a nifty (and hackable) three-channel EEG accessory for $99 — and various Android and iPhone heart-rate monitors that rely only on the phone’s camera are interesting if not entirely ready for prime time.
The interesting property of Uzma and David’s diagnostic is that the enabling technology is really dirt simple both in terms of the hardware required to make the necessary measurements and in terms of the minimal hardware calibration and the protocol for collecting the measurements. The software for analyzing the data — this is the core intellectual property — is not computationally expensive and hence the analysis can be carried out on a cell phone or performed in the cloud thus providing an additional level of security and reliability. This combination of readily available hardware and simple user interface is ideal for personal-health products and I’ll bet there are other low-hanging-fruit out there to be picked. Uzma sent us a slide deck and went through it with Greg Corrado and I after we got legal to negotiate and execuate an NDA with NYU.
One strategy for identifying such products might be to enumerate existing enabling-technology components such as blink detection, electromyography, galvanic skin response and gaze tracking, and then consult ER physicians and experienced general practitioners for ideas concerning their most useful diagnostics. I like the Google X strategy of mining catalogs for new sensors that OEMs are excited about and are likely to be available for pennies apiece in large lots. One non-obvious class of diagnostic targets might be those that require collecting data from multiple sensors over time, establishing a baseline and then looking for correlations and significant deviations from the baseline. For example, patients who don’t get much exercise often experience dizziness when getting up suddenly and this obvious symptom is often correlated with less obvious symptoms involving fluctuations in blood pressure, pulse rate and even skin temperature and conductance. Some combinations of these symptoms can be warning signs for an impending heart attack or stroke or provide early warning signs that might warrant seeing a specialist or serve to motivate behavioral changes.
December 23, 2012
Thanks again for letting me read your class notes. I picked up a bunch of useful research leads as well some ideas for my class from scanning the materials. Prompted by students I wrote up an extended “transcript” of my recent Stanford talk and put it online. It also includes some references and expanded-content footnotes and will likely serve as the introductory lecture for my class at Stanford in the Spring. I’m now working my way through Nanoscale Communication Networks by Stephen Bush [26] since it seems clear from my investigations and discussions with Greg Corrado and Fernando Pereira that reading out rather than sensing and encoding state will be the most complicated part of the puzzle.
ESB: I agree! Just ordered a copy of this book myself.
Here’s some other news briefly — I’ll be glad to fill in any details if you’re interested:
We had some exchanges with Terry Sejnowski about the Brain Activity Mapping Project that George Church and colleagues are pushing [6]. I assume you know about Terry’s activities in Washington including discussions with NIH, NSF, OSTP, DoE, DARPA and congress, the positive reception — enthusiastic in his telling — that he received regarding a big-science project along the lines of BAM.
ESB: That is good news! It sounds like the technologies we are creating, could be extremely helpful for such an endeavor. Excited to hear that people are interested.
George contacted my colleague at Google Chris Uhlik who is excited about the prospects for the technologies George is spinning but skeptical about some of the fiddly bits. Fernando Pereira is also guardedly enthusiastic. The attention is nice but everyone wants to understand whether there will be interesting projects for Google to contribute to, invest in, etc. that have relatively near-term consequences.
ESB: Yes, near-term consequences seem to always be the question! I always mention two:
detecting side effects for new neuropharmaeuticals, and
finding better brain targets for treating brain disorders.
Those are big ideas, and we are seeking to start companies and also to get innovations out into existing companies to rapidly accelerate the field of brain therapeutics.
Terry is planning a meeting at Caltech in January to discuss big-data and big-compute issues and I’ve agreed to participate representing — some dimension of — Google’s interests. Meanwhile my team is trying to figure out whether we should allocate any Q1 resources to thinking about or actually doing something about this constellation of opportunities.
ESB: Very cool! Eager to hear how the meeting goes. I participated in a meeting in DC back in December on the actual technologies, which was interesting.
In addition to puzzling through the Stephen Bush book, I’m trying to find a local electrical engineer savvy about communication networks, preferably with some nanotechnology chops who can help me with some back-of-the-envelope calculations concerning bandwidth, collateral radiation of neural tissues and low-power, short-range signal transmission.
December 21, 2012
A couple of pedagogical ideas for then next instantiation of CS379C in the Spring:
What do the economics of building a Google- or Amazon-scale data center have in common with the natural selection of more powerful primate brains and the challenge of reading off the neural state of large collections of neurons in an awake, fully functioning animal?
Think about using this Mount Everest pano and short clips from the Microcosmos film as a dramatic demonstration of scaling.
I heard an interesting story on NPR about Edward Land and the invention of the Polaroid “Instant” camera. Land started the Polaroid Corporation to develop polarizing filters for the military. The company was quite successful but Land was concerned that they needed to branch out to the consumer market to sustain their growth. He was on vacation with his daughter in 1943 and when he took her picture she asked to see it. He tried to explain that it took time to develop the film and it required some complicated chemical processes that had to be done in a lab somewhere. This got him thinking about whether this was really true. The story is that he spent the next three hours or so figuring out how to build the “Instant” technology and the next thirty years — the first consumer version of the camera wasn’t available until 1972 — perfecting the technology and making it practical. The evening that he conceived of the idea, he sat down with his patent lawyer and dictated a bunch of patent disclosures relating to the new camera. Polaroid was always quick to patent their technologies and the company benefited enormously from licensing their intellectual property especially during lean years.
December 20, 2012
Ran across this useful graphic which compactly illustrates the structural characteristics of the family of nucleotides, highlighting the phosphate groups that assist in providing the energy required for reactions catalyzed by enzymes:
 |
Notes from a conversation with Terry Sejnowski, December 19, 2012:
Terry told us — Fernando Pereira, Greg Corrado and I were on the conference call — about a meeting in Cambridge (UK) that included astrophysicists, neuroscientists and engineers and scientists working various aspects of nanotechnology. Some of them came away thinking that we are on the cusp of figuring out how to record from living brains at brain scales — billions of neurons at a time. In the US, scientists approached NSF, NIH, OSTP, DoE, and DARPA about a large-scale initiative and were enthusiastically encouraged — the Alivisatos et al [6] paper was either a prolog or postscript to these conversations.
I asked if EM-based slice-and-dice connectomics would be made irrelevant by such an effort and responded that he believed the Human Connectome Project would still be an important component in a portfolio of projects designed to accelerate progress in the field. The remainder of the conversation — right up until Terry was walking down the boarding ramp to catch his plane — touched on a bunch of scientific issues but concluded with Terry talking about how he thought companies like Google and Microsoft might contribute to the effort by providing big-data-analysis expertise.
Fernando, Greg and I talked about the prospects for two different options for reading off neural state at scale. First, could we build nanotechnology implementing tiny radio transmitters that would reside in neurons or very close to neurons in the extracellular cerebrospinal fluid. One can almost ignore what the specific technology would look like and just consider the energy required to convey the necessary information. What frequency in the electromagnetic spectrum? Note that even though the distances are small — a couple of centimeters to transmit data from inside to outside the skull — there would be a lot of transmitters working at once and contention if they were all at the same frequency, and so we would have to use clever coding and spread-spectrum techniques all of which could increase the energy requirements.
Thinking about reading off the brain in metabolic terms: Where does the energy come from? If it originates from the cell, it is important to realize that brains are perennially energy starved and so if energy is diverted to nanoscale radio transmitters then it isn’t available for other work. In addition, thermodynamics dictates that energy utilization is never perfect and our nanoscale devices will generate heat that has to be dissipated. The radio transmissions constitute a form of radiation and thus could damage a cell over time especially over longer periods of exposure.
The second option involves the idea of creating packets of data within the cell, dumping them into the cerebrospinal fluid marked for disposal and subsequently retrieving them outside of the CNS, and reading off the data. Sensing and building packets able to survive the trip might be done by re-purposing existing cellular machinery but these operations require energy and so once again metabolic economics play an important role. If we divert energy to such tasks, we may end up substantially altering the processes that we set out to study in the first place.
What about the idea of using optogenetics techniques and exploiting the fact that brains are largely transparent to bathe neurons with light and read off data from activated markers? There are energy and radiation issues here as well but usually the first object is that this option would require exposing the brain. Greg suggested that it might be worth trying to read data from transparent experimental animals like zebra fish and juvenile jumping spiders which are already the subjects of considerable interest to neuroscientists about which — particularly the developing zebrafish — we know quite a bit.
Greg provided an argument that two-photon imaging is relatively benign due to the fact that only at the focal point where two photons meet is there really enough energy to do much cellular damage. Two-photon excitation can be a superior alternative to confocal microscopy due to its deeper tissue penetration, efficient light detection and reduced phototoxicity. He also mentioned methods being developed at Max Planck Institution for several years now that were largely eclipsed by optogenetics but are still promising. A quick scan of the MPI web pages yielded a number of imaging techniques with application to neuroscience.
Notes from a conversation with Dean Gaudet, December 19, 2012:
Engineers who build industrial data centers talk about total cost of ownership. What do they mean and how does this go about driving design and ultimately the company that depends on such computational resources? These notes are partly an opportunity for me think about the idea of including a chapter on this topic in a book I’m considering. The working title for the book is “Scaling Up, Scaling Down” in admittedly obscure homage to David Kahneman’s “Thinking Fast, and Slow”, and would also include chapters on genomics, web searching, neuroscience and a bunch of other topics which I believe I can tie together in an interesting manner.
Scaling industrial data centers: Locality and the focus on data. Data has enormous value not just because people are interested in the data but because having lots of data makes it possible to draw inferences from that data, inferences that can make that data better organized and thus more efficiently accessible, better connected to other data, and ultimately more useful. Think lots of speech data allowing you to learn a better speech model, lots of financial data allowing you to connect the dots and relate economic indicators to better predict stock prices, climate data from the atmosphere, oceans, biosphere to predict the weather more accurately.
Some computationally challenged writers have commented that the giant datacenters run by Amazon, Google and Microsoft can’t approach the computational capacity of the brain. This demonstrates a lack of insight — or imagination — in understanding what computation is. In terms of work directly tied to recognizable informational products it is true that a human brain performs prodigious computational feats, but then so does almost any three-pound lump of living matter. We tend to dismiss the computations carried out by transistors at a quantum level or even at a gate-level of description. And most people have no appreciation for computations carried out in the hardware for such operations as branch-prediction, thinking only of the software level and disparaging the fact that so much of what is done in datacenters is so mundane, but then that’s doubly true of what goes on neural circuits.
Search latency, the time between when a user types a query and receives a screen full of relevant pages, is very important to users — we have very limited patience with machines. Computational latency is limited by the speed of light and the mix of accessing data and performing computations on that data. Refer to Jeff Dean’s list of numbers every engineer should know to describe differences in scale from cache references to sending a packet from Mountain View to Amsterdam and back. Not all computation is created equal.
Von Neumann machines are general purpose but they are optimized — by engineers responding to the demands of users in an interesting cycle not unlike natural selection — to expedite a particular generalized working set — a mixture of different kinds of computation in a particular distribution. What would you guess that most computation world wide is actually doing? The answer is shuffling and organizing data.
In a relatively short time, GPUs have gone from primarily a consumer product for fancy graphics and games to an essential component in the fastest computers in the world. Why is this the case, what is the mixture of computing tasks that would justify this change, and what are the consequences in terms of energy costs, computational throughput, and user latency? The Titan supercomputer at Oak Ridge National Labs and developed by Cray combines conventional CPUs with GPUs in a 1:1 ratio. Why not, say, 1:4? What is the topology of the Titan?
How do companies like Amazon, Google or Microsoft that have very different working sets than Oak Ridge think about such things? Profiling the computations performed in your data centers. Amdalh’s Law and the potential for accelerating computation by parallelization. Trading space for time. Hashing. Why so few computer scientists have any practical experience or intuitions regarding the application of hashing techniques? Trading precision for time. Single versus double precision. Thinking out of the box to reduce latency and increase user satisfaction with Google Instant. The customer at Starbucks doesn’t notice time passing if the barista keeps her occupied with conversation.
Economies of scale and bulk power rates: Energy costs. Power for components such processors and power to cool them. Heat dissipation and the environmental consequences of “dumping” heat into the oceans or atmosphere. Low power conventional CPUs and special-purpose processors like GPUs, ASICs and FPGAs. How energy is packaged and sold? Cost of transmission as a function of the distance in the power grid from generating plants. Peak time usage in a data center versus average? Why you can’t spin up a turbine at the drop of a hat.
Economies of scale and purchasing electronic components in large lots: The economics of fabricating, re-packaging, and purchasing CPU chips, circuit boards and motherboards in particular, memory and the computational costs associated with different levels in the memory hierarchy. From cache to disk and back. Technology ecosystems, multiple vendors — Intel AMD, ARM, being big enough that your purchasing power impacts what chips are good at. The advantages of commodity computing hardware over the last decade — high-performance CPUs in PCs and GPUs in gaming consoles good are examples of high-end users indirectly subsidizing the casual laptop user — and why the landscape is changing. Circuit boards at scale. Encouraging competition and innovation — to do any less is hurt yourself and the industry.
Sending a proprietary circuit design to fabricator or issuing a patent to protect your IP can be damaging whether there is theft of not. It’s the network stupid! Network fabric. The cost of moving data across a datacenter or across an ocean. Network Interface Controller (NIC) cards. Infiniband. Remote Procedure Call (RPC) and parallelism involving racks of servers. Fiber optics and the bottleneck in the electrical components. The development of optical switches fueled by DoD fear of EMP damage to critical command-and-control communication lines.
Email message sent to Chris Uhlik, December 20, 2012:
Here’s a back-of-the-envelope calculation that you might be able to carry out easily or know someone else at Google who could. Suppose you want to place a nanoscale transmitter — ultimately the gamers would like to transmit and receive but scientists are currently most interested in recording what’s going on — either inside or within a few nanometers of nearly every neuron in the primate cortex, that’s about 10 billion give or take an order of magnitude. Each transmitter would have to transmit something on the order of 40K bits per second78. The transmission distance is a few centimeters since the receiver can be positioned close to if not fitted on or attached to the skull. Transmission is through a centimeter or so of cerebrospinal fluid which is mostly just water and then layers of skin, bone, and the three layers of the meninges which enclose the cells of the brain and their fluid medium. We’d need a best guess for a low-power-radio-transmission technology that meets the bandwidth requirements. Power matters not just because the cells have to provide the power from their limited metabolic resources — neurons are always metabolically challenged — but also to minimize damage to cells and interference with their normal operation — or abnormal operation if we’re studying a brain in a disease state. I know very little about cellular or blue-tooth technologies and the problem has potential transmission interference problems that I suspect are well outside those faced by any existing technologies, and so obviously the best we can hope for here is an educated guess or extrapolation from existing technologies.
December 19, 2012
Here’s a note I sent to Steven Smith at Stanford in an exchange about the Brain Activity Map Project described in the Neuron paper [6]:
Here’s an interesting thought experiment for you. Suppose we could isolate a sample of brain tissue in vitro, read off the entire protein I/O in something approaching real-time for at least one target neuron, and simultaneously record the action potentials for that same target. I know this is currently impossible but just imagine it could be done. We’d then have a set of discrete input/output events of the form: (output) the target neuron n emitted a packet of neurotransmitter x from axon terminus r at time t, or (input) the ligand-(neurotranmitter x)-gated ion-channel c at synapse s at an unknown location on the target neuron’s dendritic arbor opened at time t and remained open for a period of δ milliseconds. (I’m just making this up as I write it and I expect you can posit a more reasonable set of events classes.) In addition, we have spike timing data for the target neuron. The first question is “Could we infer an input-output automaton that could predict the output of this neuron from a sequence of inputs?” and the second question is “Could we predict the action potentials from just the protein exchange information?” Thoughts?
And then later realizing that I hadn’t responded to one of his earlier messages I wrote the following:
I just read your recent paper [121] in Nature Reviews Neuroscience which you attached to your last email message. I knew some of the material from your Utah talk and reading an earlier draft of this paper, but the comment about synapses being “highly plastic in structure and function, influenced strongly by their own histories of impulse traffic and by signals from nearby cells” struck home more deeply in light of the gedanken experiment in the email message I sent you earlier today. The box entitled “The molecular complexity of the CNS glutamatergic synapses” made me want to cry, whether with awe or despair at the daunting prospect of modeling such an entity I’m not quite sure. Learning a static algorithm from input/output pairs is hard enough, but learning an adaptive algorithm which learns from its inputs is considerably more challenging.An interesting question from an experimental neuroscience perspective is whether or not we could somehow fix the input to the target neuron perhaps by controlling the input using, say, some method of optogenetic intervention and then collect data on the input/output behavior of the target in this artificial, but at least static environment. It might also be possible to simply “run” the target until it’s input/output pairings are observed to be stationary and then collect data for learning the now static algorithm producing that stationary behavior. Of course, the neuron will only exhibit pairings consistent with that stationary environment which may not be very interesting.
An interesting question from a machine-learning perspective is whether we have enough to go on to define a family of functions that includes functions that could explain the more complex adaptive algorithm. This sort of problem hasn’t been well studied except for very simple variants. For the most part, machine learning assumes a stationary environment. One interesting exception involves learning algorithms to implement bidding policies for automated trading agents that compete in competitive auctions. These algorithms generally employ simple models — typically variants of tit-for-tat — to predict the behavior of the other agents participating in a given auction.
I walked down the hall to talk with Aranyak Mehta, a colleague who develops algorithms for Google auctions, and we briefly discussed the possibility of modeling a neuron as a loosely coupled collection of agents, each one defined by a relatively simple algorithm involving only a very few adjustable parameters. Imagine a down-stream synaptic agent that makes a simple decision, yes or no, whether to increase or decrease the number of ligand-gated ion channels for a given neurotransmitter based on the recent history of its post-synaptic counterpart. Now the model for the entire neuron would be described by a collection of these synaptic automata plus some (possibly) centralized control that keeps track of longer-term state. Certainly such an algorithm is much more expressive than the traditional Hodgkin-Huxley family of compartmental models, but it remains to be seen if the protein-transfer-level description allows us to abstract away from the ion-concentration-level of description.
December 17, 2012
At some point, I’d like to continue with the discussion started in the December 15th entry to explore the consequences of the biological computational tradeoffs mentioned in that discussion with respect to the volatility of short-term memory as it relates to storing state information, exploiting state in the environment in the spirit of Gibson’s affordances, decision policies as they relate to the complexity of the Bellman equations, reinforcement learning and Markov decision processes, and how these relate to different types of state machines less powerful than Turing machines including push-down automata, circuit depth and current theories of executive function in primate brains, but a different question prompted this posting:
How does a signalling protein get into a cell and cause the cascade of events that results in stretch of DNA being transcribed into RNA and that RNA subsequently being translated into a protein? We have generally covered how transcription and translation work79. I’m curious about the fiddly bits such as how the cytoplasm gets suitably preloaded with all the necessary molecules — amino acids, nucleotides, phosphates, enzymes — required to transcribe DNA and translate RNA into protein. How are the concentrations of these molecules controlled locally and globally within the cell and what forces serve to bring the components together so that the required bonds are broken and formed in a timely manner? To answer this question one needs to consider existing options for the cellular transport of molecules.
Gene products including enzymes and amino acids are transported from their place of manufacture to where they are to be used by diverse cellular transport mechanisms. Previously we looked at how kinesins move along microtubule transport roots carrying molecular building blocks from the soma to locations throughout the cell. However, a lot of the intracellular (within an individual cell) and intercellular (between cells) transport and distribution that goes in the brain relies on molecular diffusion which is essentially the redistribution of liquid or gas particles within a solution due to thermal motion80. The particle diffusion equation was first described by Adolf Fick in 1855 and figured in the Hodgkin Huxley equations describing how action potentials in neurons are initiated and propagated within the axonal compartment of the cells81. Diffusion may seem like a slow and inefficient method of transfer but it is actually quite efficient from a thermodynamics perspective and it is relatively fast given the distances and temperatures involved in most neural processes.
In many cases diffusion through the cytoplasm and across semipermeable membranes accounts for how molecules move across intercellular and intracellular distances, but diffusion doesn’t account for the fine-grained adjustments of distance and molecular alignment that are needed to initiate chemical reaction. Consider the case of enzymes which are large organic molecules responsible for most of the chemical interconversions that occurs in cells. Most enzymes are proteins that act as highly specific catalysts to accelerate the rate of reactions. To be of any use, enzymes and the molecules they operate on have to be in close proximity.
Van der Waals force defined as “the sum of the attractive or repulsive forces between molecules (or between parts of the same molecule) other than those due to covalent bonds, the hydrogen bonds, or the electrostatic interaction of ions with one another or with neutral molecules.” In cellular reactions involving amino acids and phospholipids and protein-ligand interactions, the hydrogen-bonding properties of the polar hydroxyl group — which, for example, link the complementary strands of DNA that comprise the double helix — dominate the weaker van der Waals interactions. Van der Waals forces are also weak compared to covalent bonds but they don’t have to do the work of breaking or making covalent bonds, only help to position the enzymes close enough to initiate reactions in which the molecules of interest will exchange electrons to make or break covalent bonds as required82.
December 16, 2012
We’ll be talking with Terry Sejnowski on Wednesday and so I thought it might be useful to watch some of his recent talks. Terry also pointed us to paper [6] in Neuron co-authored by group of prominent neuroscientists in which they motivate the agenda for the Brain Activity Project which Terry is promoting. Here is the link to a recent talk by Terry in March of 2012:
Brains need to make quick sense of massive amounts of ambiguous information with minimal energy costs and have evolved an intriguing mixture of analog and digital mechanisms to allow this efficiency. Analog electrical and biochemical signals inside neurons are used for integrating synaptic inputs from other neurons. The digital part is the all-or-none action potential, or spike, that lasts for a millisecond or less and is used to send messages over a long distance. Spike coincidences occur when neurons fire together at nearly the same time. In this lecture I will show how rare spike coincidences can be used efficiently to represent important visual events and how this architecture can be implemented with analog VLSI technology to simplify the early stages of visual processing.Here is the link to another talk, this time at the Allen Institute in October of 2011:
Dr. Sejnowski began with a computer-animated tour of the hippocampus, reconstructing a small cube, measuring micrometers on a side, of intra- and extra-cellular space at a resolution of 4nm. Zooming in so closely on synapses of the neuropil – the space between neuronal cell bodies where axons and dendrites come together – led to a marveling at the effectiveness of synapses. Great detailed analysis can be performed at this level of magnitude, and Dr. Sejnowski pointed out just a few important observations. Extracellular space is not uniform. The spines and heads of dendrites have enormous space and size variation, and post-synaptic density is directly and linearly related to spiny head volume. What this means is an open question in neuroscience.Finally, here is the link to Sejnowski’s talk at the Redwood Institute on October of 2005.
December 15, 2012
When nature got around to building bigger brains it already had all the machinery for scaling the basic plumbing in place. Transporting and distributing nutrients to billions of cells had to be figured out millions of years of ago. Ditto for waste management and communication over body-spanning distances. Parallel computations of an embarrassingly parallel sort was the norm but there were good reasons for centralized processing and natural selection made some reasonable trade-offs. Of course, there were computations that it couldn’t perform in parallel and so there were compromises made to trade space for time to avoid the antelope getting slagged while contemplating a conditional branch in its decision tree. Logic didn’t fit neatly within the existing technology in large part due to the need to keep information on the stack and brains hadn’t much use for a deep stack; either you stored it in global memory keyed to an appropriate stimulus or you kept it in memory by some form of rehearsal. It wasn’t that nature didn’t have machinery for keeping things in short-term memory, it was just that the machinery wasn’t easy to adapt to conscious serial-thinking.
Brains are good at the sort of things GPUs do well; lots of parallel threads running more or less independently. Streaming processors (SP) in GPU devices and SIMD units in traditional CPU cores can’t handle conditionals except by sequentially masking all bot one path through the conditional. Traditional CPU cores are very good at conditionals and even have special purpose hardware to predict the most likely branch. For some applications you’d like lots of cores all of which are good at branching but you pay a price in terms of real estate on the silicon die dedicated to branch prediction and other optimizations that accelerated largely serial code. If you can control branching, say, by packaging all the threads that branch one way to execute in one SP and all the threads that branch the other way to run in a different SP, then you can take advantage of a GPU with multiple SPs. Brains are mostly like GPUs in that they can do lots of things in parallel as long as the different tasks don’t require coordination and don’t do a lot of deep branching with recursive procedure calls which requires keeping a stack to you can backtrack to previous branch points. Traditional von Neumann architecture CPUs are optimized for branching but they need a lot of extra hardware to keep track of local state and as a result you can’t get as many of them on a single silicon die.
December 14, 2012
An explanation of why Google Brain models with enormous numbers of parameters don’t get caught in local minima — or at least local minima with poor performance, and why they don’t overfit the data — or at least if they do overfit then it doesn’t substantially reduce performance. The more parameters the larger the set of models being explored but also the more dimensions to work with in searching for a good model and the more models that are likely to be perform reasonably well. The flip side is that though there may be more models that perform reasonable well, these still may be buried in a much larger set of models that perform poorly and hence just as hard to find. It could be that as the total number of models increases the fraction of models that exhibit good performance actually increases or perhaps the fraction remains constant but, since the parameters are highly redundant it is easier to find a good model by stochastic gradient descent. As for overfitting, we generally restrict the expressive power of our models because we believe that simpler models are more likely to generalize. However, if we have enough training data such that we’ve really covered the space of possible input, then it may not be a mistake to memorize the training data as long as there is the models have some built in tolerance for noise and small perturbations in the input representations.
December 12, 2012
Discussion with Jay about how to turn an interesting scientific opportunity into a viable project at Google. How to think about crafting a three-sentence elevator pitch?
Summary: Figure out milestones and near-term deliverables that provide one or more of the following: substantial and sustainable revenue streams, enabling technologies that provide a strategic technology advantage in a relatively short time horizon — think Google Glass, or a significant impact on society that we are uniquely positioned to make happen and that would be impossible or substantially delayed without our intervention — think self-driving cars.
Homework: How soon until we can read off the connectome of a fly? A mouse? A monkey? How soon until we can do so without harming the experimental subject? How soon can we read off the time-sampled proteome in vivo in something approaching real time and continue doing so indefinitely? How soon until we can complete the loop using a (relatively) non-invasive technology such as optogenetic sensing to provide feedback to the user?
Homework: At what point would the medical applications of this technology likely yield clinical treatments for Alzheimer’s, Huntington’s, Parkinson’s, or other some other relatively common neurodegenerative disease? Besides developing technology to collect data for basic science, might there be personal-health technologies, e.g., monitoring the brains of stroke victims and the elderly or children at risk for autism and other developmental disorders — that might have a more immediate impact.
Case study: Sebastian Thrun’s project to hire robotics and control-systems engineers to develop self-driving cars. What’s the upside for Google? Perhaps it’s the combination of positive publicity, substantial impact in terms of lower energy use and environmental impact, reduced loss of life due to accident and pushing the state of the art while leveraging our mapping data for the greater good.
December 11, 2012
Fernando sent me a whitepaper written by Terry Sejnowski with the title “The ‘Brain Activity Map’ Grand Challenge”. Terry’s outline covers the programs that we’ve been encouraging and funding — Sebastian’s connectomics and Ed’s robotic-patch clamping, 3-D recording arrays and optogenetics — in his list of short term (5 year) goals, and the new stuff I’ve been peddling in talks at Stanford and Brown and discussing with Boyden and Church — the application of synthetic biology, recombinant DNA technology and hybrid nanotechnology to scaling neural readout — in his longer term goals.
I told Fernando that “[t]his is something that Google ought to push and become a part of — it’s going to happen, probably faster than we think, Google can help to accelerate it, there is an important role for big data / massive compute and fast signal processing, we should be playing in the area of synthetic biology and currently we haven’t a clue and hardly any engineers with expertise (this despite how popular synthetic bioengineering classes are at the top engineering schools), and, if that wasn’t enough, the area is so exciting and challenging that it will draw the smartest, most ambitious engineers like a magnet and we can’t afford to lose out on attracting them to work at Google.”
December 10, 2012
Church and Regis [36] give a nice account of the origins of bioengineering and engineered biological components. They tell about Tom Knight’s crusade to build reliable, standardized biological components and his seminal paper entitled “Idempotent Vector Design for Standard Assembly of Biobricks”. They recount the saga of Drew Endy’s synthetic biology course which was offered at MIT in 2003 with the title “Synthetic Biology: Engineered Genetic Blinkers”, and the fact that none of the parts resulting from student projects worked as advertised, and only a few could actually be sequenced. The course was repeated in 2004 with roughly the same outcome.
Then came iGEM (intercollegiate Genetically Engineered Machines) which was held in the Summer of 2004 and kicked off with the challenge of building biological finite state machines. The competition ended up with several successful projects including an entry from UT Austin that created an E. coli film that could take the biological equivalent of a photo. As a demonstration they created a film that displayed “Hello World” in homage to Brian Kernighan’s first program for beginners learning the C programming language.
Knight and Endy’s dream of building a registry of standard biological parts is now a reality with a catalog including over 10,000 standard components and suppliers that provide these components at competitive prices. IGEM — with the “intercollegiate” changed to “International” — is now so large it is held at several locations worldwide and the winning entries include incredible projects ranging from high-density biological data storage systems to a strain of yeast designed to terraform the planet Mars. There is also grass-roots movement to make molecular engineering something that anyone can learn about and experiment with.
Ed Boyden’s current MIT course Neurotechnology Ventures offers yet another outlet for creative application of synthetic biology, namely the nourishing of the entrepreneurial spirit. The listing for the course includes this description:
Special seminar focusing on the challenges of envisioning, planning, and building start-ups that are commercializing innovations from neuroscience and the blossoming domain of neuroengineering. Topics include neuroimaging and diagnostics, psychophysiology, rehab feedback, affective computing, neurotherapeutics, surgical tools, neuropharmaceuticals, deep brain stimulation, prosthetics and neurobionics, artificial senses, nerve regeneration, and more. Each class is devoted to a specific topic area. The first hour covers the topic in survey form. The second hour is dedicated to a live case study of a specific organization. A broad spectrum of issues, from the deeply technical through market opportunity, is explored in each class.On a whim, I typed in go/magnet and ran a bunch of exploratory queries. The query “neuroscience” nets eleven people most of whom appear to have only passing knowledge of the subject and the rest I’ve met or know of. No one answers persuasively to any one of the queries: “molecular engineering”, “synthetic biology”, “bioengineering”, “genetic engineering”, “recombinant DNA technology” or reasonable combinations and permutations thereof. Presumably Tim Blakely, whom I mentioned in a previous post, would have come up had he filled in a profile in magnet but perhaps interns are not encouraged to register.
December 9, 2012
Zador et al [172] have proposed the idea of sequencing the connectome in a paper of the same title. The authors break down the problem into three components: (a) label each neuron with a unique DNA sequence or barcode — a discussion of recent work on DNA barcoding follows, (b) propagate the barcodes from each source neuron to each synaptically-adjacent sink neuron — this results in each neuron collecting a “bag of barcodes”, and (c) for each neuron combine its barcodes in source-sink pairs for subsequent high-throughput sequencing.
It would seem the authors have in mind sacrificing the animal in the final step, but it may be possible to pass source-sink pairs through the cell membrane and into the lymph-blood system for external harvest as proposed in an earlier post. They suggest that propagation might be accomplished via some variation using a trans-synaptic virus such as rabies [122] or some extension existing brainbow technology [57] both of which we’ve talked about. Additional techniques would be required to map barcodes to brain areas. The authors claim to be developing an approach to based on PhiC31 integrase for joining barcodes and PRV amplicons [126] for trans-synapatic barcode propogation.
George Church and his colleagues at Harvard have developed a method of fluorescent barcoding to tag and subsequently identify DNA [97].83 Their work relies on building robust substrates out of DNA origami84 which was developed by Paul Rothemund [130] at Caltech and has shown promise as a programmable drug delivery mechanism. Ed Boyden has mentioned the possibility of collaborating with the Church lab to apply DNA barcoding techniques to sequencing the connectome.
Church and science writer Ed Regis recently released a popular-science book on synthetic biology and the burgeoning biotech industry [36]. I haven’t finished reading the book, but so-far I’ve picked up some interesting factoids such as how Craig Venter while at NIH took advantage of the fact that they had lots of money but little space for equipment and so he out-sourced almost everything and used complementary DNA (cDNA) with the introns excised to simplify the sequencing.
I sent the following note to Ed this morning:
I was misled in searching for “DNA barcodes” by related work on using existing DNA for taxonomic purposes. Once I looked at the Church lab pubs pages I discovered what must have interested you in collaborating with them. In addition, I re-read the 2012 Zador et al [172] paper in detail following up on several of the technologies they propose for sequencing connectomes. It is a simpler problem than I originally looked at, but more reasonable as an interim goal. Even in this case, there are lots of moving parts and existing methods for generating barcodes and propagating them across synapses with retroviruses will have be adapted. I assume they have in mind doing the analysis in vitro or in vivo and subsequently sacrificing the animal to do the sequencing. What’s your prediction for how long it will take someone to develop the technology to successfully sequence a connectome using some variant of the idea described in the Zador et al paper? Two years? Five?He answered as follows — in his remarks, “Tony” is Anthony Zador at Cold Spring Harbor Laboratory and “Koerding” is Konrad Koërding at Northwestern:
Yes — in addition, Church, us, and Koerding all came up with the idea independently at the same time, and decided to work together rather than compete. So it’s also part of this new model of collaborating on neurotechnology given how difficult it is. I think that some variant of the idea could be relatively quickly realized. I know Tony is pushing hard on multiple versions of the idea. At least 2–3 other groups seem to be working on related ideas, if not more. We are collaborating with Tony on the ability to utilize the technology also to understand what cell types are connected, since a completely anonymous connectome (e.g., not knowing what kinds of cell are being connected) might only be of minor interest.When I asked about funding he said that Tony already has funding from both Paul Allen and NIH to pursue this line of research. Tony worked with Christof Koch at Caltech and Christof is now the Chief Scientific Officer at the Allen Institute for Brain Science.
If you missed the Exascale “Early Results for Computation Based Discovery” talks, you might want to check out Tim Blakely’s open-source BigBrain Project. Tim is an intern working with Joey Hellerstein and 6th year PhD student in Bioengineering at the University of Washington studying Brain-Computer Interfaces.
December 8, 2012
Here’s a cheat sheet of shortcuts to Wikipedia entries relevant to our discussion of organic chemistry:
A polypeptide is a short polymer consisting of “amino acid monomers linked by peptide bonds, the covalent chemical bonds formed between two molecules when the carboxyl group of one molecule reacts with the amino group of the other molecule. Peptides are distinguished from proteins on the basis of size, typically containing fewer than 50 monomer units.” A polymer is just a chemical compound consisting of repeating structural units.
The lipids constitute a “broad group of naturally occurring molecules that include fats, waxes, sterols, fat-soluble vitamins (such as vitamins A, D, E, and K), monoglycerides, diglycerides, triglycerides, phospholipids, and others. The main biological functions of lipids include energy storage, as structural components of cell membranes, and as important signaling molecules.”
A sugar is special case of a carbohydrate and, as this name implies, they “are composed of carbon, hydrogen and oxygen, usually with a hydrogen:oxygen atom ratio of 2:1 (as in water); in other words, with the empirical formula Cm(H2O)n.” Deoxyribose — a component of DNA — is a notable exception and has the empirical formula C5H10O4. Sugars play an important structural role in the genomic pathways we have been looking at, e.g., the “5-carbon monosaccharide ribose is an important component of coenzymes (e.g., ATP, FAD, and NAD) and the backbone of the genetic molecule known as RNA.”
Adenosine-5’-triphosphate (ATP) is a “nucleoside triphosphate used in cells as a coenzyme. It is often called the ‘molecular unit of currency’ of intracellular energy transfer. [...] Metabolic processes that use ATP as an energy source convert it back into its precursors. ATP is therefore continuously recycled in organisms: the human body, which on average contains only 250 grams (8.8 oz) of ATP, turns over its own body weight equivalent in ATP each day.” When ATP is used in a reaction it must be reconstituted by the following reaction: ADP + Pi yields ATP, where ADP (adenosine diphosphate) and the phosphate group Pi are joined together by the enzyme ATPsynthase.
December 7, 2012
I was watching Eric Lander’s video describing PCR (Lecture 18 of the OpenCourseWare Introduction to Biology). It was 2004 and the Red Sox had just won the league championship coming back from three-game deficit and were preparing for the World Series. They went on to sweep the Cardinals and win their first World Series since 1918 (86 years). The Human Genome paper with its 2,000 some authors including Lander came out on the day of the lecture and it was the week before the election in which George W. Bush defeated John Kerry, then junior democratic sensor from Massachusetts, to win his second term. Pretty heady times for an MIT freshman.
Lecture 18 also includes a nice discussion of shotgun sequencing which Fred Sanger developed around 1980. The previous lecture (17) described various methods for sequencing shorter sequences emphasizing variations on the method developed by Sanger and his colleagues which first saw use around 1977. This technology was combined with the shotgun method by Human Genome Project (HGP) which succeeded in reading the human genome85 in 2003, but is rapidly being supplanted by so-called next- or third-generation technologies including those from Ion Torrent, Oxford Nanopore and Complete Genomics. Variations on the Sanger method require lots of reagents to extend DNA templates from bound primers followed by heat denaturing and separation by size using using gel electrophoresis. The process can be parallelized and sped up using radioactive or fluorescent tags to support optical readout of bases.
Lander’s Lecture 17 describes basic sequencing technology including the Sanger chain-termination method. I like to have lots of options when trying understand a new idea or procedure, especially in biology and the life sciences where I can easily get overwhelmed by all the terminology. I found the treatment of the Sanger method in Chapter 23 of Molecular Biology by David Clark and Lonnie Russell easier going in large part because of the excellent graphics. Your mileage may vary, but I recommend that you ask around about the learning resources available on-line and perhaps purchase or check out of your local library a sample of molecular biology reference books, including an introductory and an intermediate text. The Clark book is now in its 4th Edition, and, for a more advanced book, I recommend From Genes to Genomes: Concepts and Applications of DNA Technology by Jeremy Dale, Malcolm von Schantz and Nick Plant.
Off the subject of DNA sequencing, I ran across an article by Jonah Lehrer in the New Yorker debunking the over-hyped benefits of brainstorming. It’s a good summary of what works and what doesn’t and a nice illustration of how a charismatic huckster can take advantage of people who want to believe something — in this case the idea that to unleash our inner creativity it is necessary to hold back on judgement and critical feedback — despite the lack of supporting evidence. Alex Osborn, the high-priest of nonjudgmental groupthink, wrote with no justfication that “[c]reativity is so delicate a flower that praise tends to make it bloom while discouragement often nips it in the bud.” Lehrer refers to several replicated studies showing that criticism improves outcomes and that working collectively is often not as effective as starting out by working independently and then subsequently getting together to refine, combine and spawn additional ideas.
December 5, 2012
Check out this article in the Economist describing recent results from the ENCODE Project 86 and conjectures concerning the previously unsuspected role of so-called junk DNA in the human genome. The article focuses on a study published in Genome Biology written by David Kelley and John Rinn of Harvard University that looks at one particular class of such genes called lincRNAS. By the way, there are now lots of great tools for exploring the ENCODE data; you can experiment with one them here.
December 4, 2012
Here’s the text of a note that I sent to Ed Boyden this morning: I found your account [22] of the development of optogenetics inspirational and instructive. I particularly liked your acknowledging the role of serendipity, luck and the confluence of researchers simultaneously and aggressively — in a good way — trying to solve the same or similar problems using the same tools and more often than not willing to share what they’ve learned in terms of tools, techniques, gene products, reagents, etc. And thanks for the pointer referring to “genetic barcodes”. Your pointer led me to the barcode work on yeast and the Church Lab at Harvard. I’m scouring the literature looking for various tools and techniques to populate my bag of tricks for thinking about proteomics-enabled connectomics, while simultaneously back filling my scattered knowledge of molecular biology.
I think I’d like taking your “Principles of Neuroengineering” and look forward to the day when you or one of your students or colleagues can find the time to put together a course like Bob Weinberg and Eric Lander’s “Introduction to Biology”, which inspired me and I suspect legions of MIT students and the many students elsewhere who’ve stumbled across the videos on the web. It’s so great working in a wired world where you can easily search for and find such great resources; if I was suddenly transported back to a time when I had to look up everything in a library and communicate with my colleagues via snail mail, I think I’d revert back to mathematics.
December 3, 2012
Learned some interesting statistics regarding coding errors during transcription. In its “initial pass”, DNA polymerase makes about 1 mistake in 1000, which essentially means that even the smallest genes will have mutations. But DNA polymerase has an additional option to remand or “subtract” an incorrectly transcribed nucleotide and, if it has made such an error, then with a probability better than chance it will appropriately perform the subtraction. This proofreading ability reduces the error rate to something closer to 1 in 104 or 105. There is also an enzyme that identifies mismatches and marks them for subsequent repair which requires some more complicated splicing enzymes. This mismatch-repair step reduces the error to around 1 in 108. In addition there are error correcting steps for primers, caps, telomeres, etc.
In principle, each three-nucleotide (messenger RNA) codon can encode any one of 43 = 64 amino acids but only 21 amino acids are used by eukaryotes and there are two stop codons employed by some organisms. There are plenty of non-coding RNA molecules found in cells — it is estimated that humans have on the order of 10,000 non-coding RNAs — including transfer RNA, ribosomal RNA micro RNA, and, it is expected, many more that may be non functional. Note that if a ribosome gets to the end of a messenger RNA and doesn’t encounter a stop codon, it generally means that it made a mistake or that the messenger RNA is ill-formed in which case additional cellular machinery tags the “stuck” ribosome for degradation — yet another mechanism for reducing errors or mitigating the consequences of earlier errors.
In Lecture 13 in Introduction to Biology, Claudette Gardel discusses “protein localization”, describing the basic mechanisms for how proteins products get in, e.g., how lactose enters the cell to trigger the production of lactase and out, e.g., how insulin produced in the cells of the pancreas exits the cell membrane. She also touches upon how proteins move across membranes other than inner and outer cell membranes, namely membranes of the nucleus, endoplasmic reticulum, Golgi apparatus and mitochondria.
Thinking about the continuity of life from single-cell organisms starting around 3B years ago, to eukaryote cells about 1.5B and the first multicellular organisms about 1B years back. The first mammals appeared around 100M years ago in the Mesozoic. It is interesting to think that each of our cells can be traced back though a long series of living organisms to these ancestor cells. Each mutation along the way conferred some reproductive advantage or, at the very least, didn’t kill the organism before it had a chance to replicate. Replication itself is extraordinarily interesting and doesn’t take much reflection to understand why the likes of John von Neumann, John Conway and Alan Turing were fascinated with self reproducing machines. I think a good exercise to get High School students excited about biology would be to have them break up in teams, try to design such a machine and critique one another to identify design flaws — it is so easy to inadvertently anthropomorphize some aspect of reproduction, get the recursion wrong or forget to include some seemingly trivial but essential computational step as part of the organism.
December 1, 2012
The Economist had a series of articles over the last year or so, all of which have the title “The Eyes Have It”, including one in the June 30 issue relating to wearable computing and featuring a photo of Sergey Brin wearing the Google Glass prototype, the article in the July 7 issue relating to biometrics, security and the Afgan war, and now the December 1 issue relating to gaze tracking. The Economist’s depiction of the state-of-the-art technology is pretty accurate if somewhat vague and their projection for rapidly falling prices is probably even less optimistic than is warranted — that is “optimism” assuming you are hoping for ubiquitous, cheap eye tracking.
November 27, 2012
I expect that many of the students in Yoav’s class were bewildered by much of what I had to say. The material in my lecture covered several disciplines and some of the points I made are subtle and take some getting used to. I suggested that Yoav challenge the students interested in learning more to conduct the following exercise: Read the abstract of the 2005 paper on optogenetics by[23] and then go straight to the “Methods” section. Read each sentence in sequence and if there’s any term you don’t understand then look it up on Wikipedia; create an appendix in anticipation of writing a short precis summarizing Boyden’s paper and containing each term you had to look up along with an abbreviated definition in your own words.
In reading the Wikipedia entries, you will likely encounter additional terms you are unfamiliar and so recurse until you feel comfortable with your understanding of the term you started with. When you’re done you’ll have a tree — or perhaps forest — of relevant terms. Along the way, take the time to try to answer questions that come up such as “Why do they work with hippocampal cells?” — if you answer this particular question you’ll learn something about the life cycle of neurons. When your appendix is reasonably complete, go back to the abstract — or to the methods section if you’re more ambitious — and rewrite the abstract in your own words expanding concepts that you wish were made clearer when you first read it. I guarantee that if you carry out this exercise you’ll learn a great deal and you’ll retain that knowledge longer and in more detail than is possible with less focused learning.
I do this sort of focused reading all the time and have discovered that not only does it improve my understanding but I find it much less frustrating, since, rather than trying to grasp some complex idea all at once, I am constantly learning bits and pieces of the larger puzzle and integrating them into the rest of what I know about the subject. It can take a fair bit of time to read a paper in a discipline you’re unfamiliar with but the method quickly pays dividends and soon you’ve compiled a glossary and set of references that you can easily navigate in and you’ll be reading and understanding new papers much more quickly. A while back, I read selected chapters in “From Genes to Genomes: Concepts and Applications of DNA Technology” by Jeremy Dale, Malcolm von Schantz and Nick Plant, and here’s a partial listing of the items that I compiled in reading the first chapter:
A plasmid is “a small DNA molecule that is physically separate from, and can replicate independently of, chromosomal DNA within a cell. [...] Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms.”
A vector is “a DNA molecule used as a vehicle to transfer foreign genetic material into another cell. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.”
A selectable marker is “a gene introduced into a cell [...] that confers a trait suitable for artificial selection. They are a type of reporter gene used [...] to indicate the success of a [...] procedure meant to introduce foreign DNA into a cell [and include] antibiotic resistance genes.”
A ligase (DNA or RNA) is “is a specific type of enzyme, a ligase, that facilitates the joining of DNA [RNA] strands together by catalyzing the formation of a phosphodiester bond. [...] It plays a role in repairing single-strand breaks in duplex DNA in living organisms.”
A polymerase chain reaction (PCR) is “an extremely versatile technique for copying DNA. [...] PCR allows a single DNA sequence to be copied (millions of times), or altered in predetermined ways.”
A restriction enzyme (or restriction endonuclease) is “an enzyme that cuts DNA at specific recognition nucleotide sequences known as restriction sites. [...] Inside a bacteria, the restriction enzymes selectively cut up foreign DNA in a process called restriction; while host DNA is protected by a modification enzyme (a methylase) that modifies the bacterial DNA and blocks cleavage.”
A bacteriophage or just phage is “a virus that infects and replicates within bacteria. [...] Bacteriophage are composed of proteins that encapsulate a DNA or RNA genome [and] replicate within bacteria following the injection of their genome into the cytoplasm.”
Following transplantation into the host, the foreign “DNA contained within the recombinant DNA construct may or may not be expressed. DNA may simply be replicated without expression, or it may be transcribed and translated so that a recombinant protein is produced [which may require] restructuring the gene to include sequences that are required for producing a mRNA molecule that can be used by the host’s translational apparatus [promoter, translational initiation signal, and transcriptional terminator].”
A nanopore is “a small hole, of the order of 1 nanometer in internal diameter. Certain porous transmembrane cellular proteins act as nanopores, and nanopores have also been made by etching a somewhat larger hole (several tens of nanometers) in a piece of silicon, and then gradually filling it in using ion-beam sculpting methods which results in a much smaller diameter hole: the nanopore. Graphene is also being explored as a synthetic substrate for solid-state nanopores.”
November 25, 2012
Since first thinking about the idea a couple of days ago, I’ve come up with a dozen or more pages of written notes, a bunch of alternative “algorithms” and a boatload of pointers to related ideas both speculative — from futurists and science fiction writers — and provocatively practical — from scientists and engineers. The slides for the talk on Monday are here, but they were primarily a convenient method of forcing me to work hard to get the facts right. The slides mainly consist of pretty pictures to keep the visually oriented alert; the content is in my head and scattered throughout these notes. Some of the slide titles need a bit of explanation since I treat them as conversational gambits to engage the students. For example, I don’t consider Sebastian Seung’s connectome work or Ed Boyden’s 3-D cell-recording and robotic patch-clamp work as scalable in an interesting way. The slides contrasting “Tiny Targets” and “Tiny Machines” are meant to drive home this point.
I continue to be excited about the prospect of developing this sort of technology and believe it would be worth the effort to search the literature carefully to identify existing technologies that might be adapted for our purpose and to think in detail about algorithms for data analysis that would scale to address the computational and storage problems that an instance of the sort of sensor technology I envision would entail. The effort may or may not yield a patent but we might be able to identify partners to handle the chemistry and molecular biology pieces of the puzzle and coordinate with Google or a yet-to-be-determined startup funded by Google Ventures to bring all the pieces together. Alright, I admit I can get carried away, but this is the most upbeat I’ve been about the near-term prospects for computational neuroscience in years.
Several science fiction writers and futurists have considered the idea of delivering nanoscale machines to the brain via the blood supply, e.g., in a 2009 interview Ray Kurzweil suggested:
By the late 2020s, nanobots in our brain (that will get there noninvasively, through the capillaries) will create full-immersion virtual-reality environments from within the nervous system. So if you want to go into virtual reality the nanobots shut down the signals coming from your real senses and replace them with the signals that your brain would be receiving if you were actually in the virtual environment. So this will provide full-immersion virtual reality incorporating all of the senses.and I expect that Kurzweil, Hans Moravec and probably quite a few other futurists and science fiction writers have spun similar stories in print since the publication of Eric Drexler’s Engines of Creation in 1986. (See the article “The Singularity is Far: A Neuroscientist’s View” for a contrarian view written by a neuroscientist.) There is also quite a bit of buzz — “techno babble” really — about using some form of nanotechnology to knock out cancer cells, overcome drug addiction and cure warts — just kidding about the last. While perhaps a detail to reporters, the current proposal is seriously considering nanoscale machines designed from scratch but rather adapting existing molecular machinery in much the same way as optogenetics uses recombinant DNA technology to adapt voltage-gated ion channels to open or close in response to a light signal.
It may also be possible to tag molecules in such a way to record biological events that would be sufficient to infer interesting global properties of neuron ensembles if only we could transmit the information to a computer for processing. There are on the order of 1014 edges (synapses) in the human cortical connectome, on the order of 1011 nodes (neurons) and, say, a few hundred neurotransmitters responsible for most of what goes on in the brain. We can encode any integer between 0 and 18,446,744,073,709,551,615 in 64 bits. In principle, we could tag each neuron, each of its synapses, and each class of neurotransmitters with a unique 64 bit integer. What if could record every event (or a representative sample of such events) of the form: neurotransmitter X originating from neuron A was received at neuron B at time T, encode the information in an RNA sequence, encapsulate it in a protective protein shell like a viral capsid, and boot it out of the CNS appropriately tagged so it can be identified and filtered out of the blood stream for analysis.
On the topic of using sequencing to infer connectomic information, Zador et al [172] have recast the study of neural circuitry as a problem of high-throughput DNA sequencing. The authors claim that such a method has the potential to increase efficiency by orders of magnitude over methods that depend on light or electron microscopy. My concern about passing retro-viruses through the blood brain barrier were not entirely unfounded but nature has figured out workarounds to deliver some of its scarier brain pathogens. The papers I found on the role of astrocytes and the endothelium suggest we might learn some tricks from these pathogens that could simplify the sensing interface.87 Speaking of nasty pathogens, check out the Wikipedia page, YouTube video and New York Times article on viral-based neuronal circuit tracing — the virus in this case is rabies. Unlike the approach that we’ve been considering in our previous posts, this method uses a self-replicating viral vector to propagate markers throughout the cell and to adjacent cells connected through axonal processes.88
November 23, 2012
How do you design a retro-virus delivery vector that only targets neurons, doesn’t replicate, achieves high coverage of the target population, and can be manufactured economically in sufficient quantity?
Could you do so in way that the encapsulated RNA instructions are identical except for a unique signature which will be used to tag the host neuron for collecting connection attributes?
If not, could you induce the host to generate such a signature exactly once upon the first being infected, and provide some guarantee that the self-manufactured signature is with high probability unique within a given population of neurons all of which use the same method for generating their signatures?
How might we introduce the vector into the blood supply, avoid an immune response rejection, circumvent the blood-brain barrier, reliably makes its way to the host, and quietly self-destruct if the host is already infected?
Could you encapsulate the vector in a cell — perhaps the virus capsid will suffice — or alternative packaging that is specifically keyed to the endothelial membrane surrounding the capillaries?
If not, perhaps the endothelial cells could be modified to admit a specially packaged retro-viruses — this warrants a literature search to look into viral pathogens that attack the central nervous system?
What are some candidate cellular machines that could be adapted to sense neural states and how might they be so modified to carry out sensing operations without interfering with their normal function?
Could you alter a ribosome so that as a side effect of transcription it also produces a marker for the particular protein — perhaps it could do this some fraction of the time such that the marked proteins are proportional to total production?
If not, could a completed protein be tagged or might it be better to tag and package the mRNA after it has performed its purpose assuming that the process leaves the single-stranded mRNA intact?
Once the data has been recorded and packaged to include the necessary provenance to enable reconstruction of the information and is either floating free in within the cell membrane or secured to some organelle as a staging area for subsequent post processing, how would you perform any additional protective cloaking and transfer the packaged information outside the cell body and into the blood / lymph system?
November 21, 2012
I promised a colleague at Stanford that I’d give a guest lecture in this class on “something having to do with brains and neuroscience.” As usual, I decided to use the opportunity to think about something new and so I set about creating a slide deck and collection of supplementary notes on an idea that I’ve been knocking around for a couple of months. The idea concerns an alternative approach to collecting data from large collections of neurons; I like to think of it as a neuroscientist’s take on Celera’s shotgun approach to genome sequencing.
I have a friend who works in the pharmaceutical industry with a focus on genomics. I gave him a quick version of my idea and he cautioned me against blurting it out to a bunch of Stanford students looking for startup ideas — he thinks all Stanford students are looking for startup ideas and given my interactions lately I don’t think he is far from the mark. I tend to have a low opinion of the novelty of my own ideas and told him I didn’t think the idea was worked out fully enough to warrant more than cursory attention, but he said he’s filed lots of patents with less novelty and far less content than the one I was pitching. Here’s the original pitch:
Methods like Sebastian Seung’s slice-scan-and-segment approach to inferring the connectome or Ed Boyden’s 3-D probes for multi-cell recording and robot-controlled patch-clamping and probe insertion won’t scale. Ditto for the more recent work on using optogenetic sensor scans for learning neural circuits.
We describe an approach for inferring connectomic and functional status of large populations of neurons by utilizing existing blood and lymph networks to induce neural targets to conduct sensing and data collection operations using their existing genomic machinery augmented by nanoscale machines introduced using recombinant DNA technology.
A non-replicating retro-virus is distributed to a target population of neurons via the capillaries that supply blood to the brain. This retro-virus is designed to circumvent the blood brain barrier by utilizing genetically modified astroscytes and endothelial cells.
The retro-viruses would carry RNA to adapt existing molecular machinery to provide sensing and coding functions in addition to their original cell functions. The adapted machinery would sense the type and expression level of proteins, encoding this information in RNA packets for subsequent forwarding to destinations outside the CNS.
Data collection timing and cell identification information would be packaged with the protein expression statistics and the resulting packets shunted back into the blood and lymph networks. Cells identifiers would consist of random RNA signatures and timing would be based on counters synchronized using phase-offset alpha and beta rhythms.
Packets released back into the blood stream would be intercepted by a cell sorter and shunted through a processor that would read out the encoded information and convey it to a device outside the organism using some form of light or radio transmission. The packet processor and transmission machinery needn’t conform to the same size and immune-rejection requirements as the instruction packets that pass through the blood brain barrier.
Additional markers could be used to infer connectomic information by tagging neurotransmitters and it may even be possible to sense voltage levels in compartments and convey this information as well. The collected data could be used to infer network structure — relevant to connectomics, signalling pathways — relevant to proteomics, evidence of pathologies — relevant to clinical practice, etc.
Your timely feedback will be most welcome. I’m not proud; if it’s obvious, too far out, been done before, or you’re just not interested, please say so and I won’t be offended. Quite the contrary you may save me some embarrassment or disappointment down the road. Thanks.
November 19, 2012
The Brainbow technology being developed at Harvard is an alternative or possibly a useful complement to the approach pursued by Sebastian Seung [136] for constructing the connectome of an organism’s brain or other neural tissues such as the retina. It is a “process by which individual neurons in the brain can be distinguished from neighboring neurons using fluorescent proteins. By randomly expressing different ratios of red, green, and blue derivatives of green fluorescent protein in individual neurons, it is possible to flag each neuron with a distinctive color. This process has been a major contribution to the field of connectomics, or the study of neural connections in the brain.” This approach has a number of drawbacks including the requirement of breeding at least two strains of transgenic animals from embryonic stem cells which is both time consuming and complex.
Optogenetics [23] is “the integration of optics and genetics to control precisely defined events within specific cells of living tissue even within freely moving animals, with the temporal precision (millisecond-timescale) needed to keep pace with functioning intact biological systems.” I mention it in the context of reading brain states because the development of optogenetic technology necessarily includes the development of “genetic targeting strategies such as cell-specific promoters or other customized conditionally-active viruses, to deliver the light-sensitive probes to specific populations of neurons in the brain of living animals, e.g. worms, fruit flies, mice, rats, and monkeys.” These same developments would serve as technology enablers for the sort of non-invasive, neural-state sensing that I’m proposing — see Bernstein and Boyden [16] for a discussion of the optogenetic toolkit.
Specifically the channelrhodopsins used in optogenetics can be readily expressed in excitable cells such as neurons using a variety of transfection techniques, e.g., viral transduction, cell-plasma-membrane electroporation. In the case of optogenetics, the channelrhodopsins function as light-gated ion channels. The so-called C-terminus of the channelrhodopsin extends into the intracellular space and can be replaced by fluorescent proteins without affecting channel function. Researchers are already developing light-controlled probes using related technology that supports sensing brain states [101].
Optogenetic technologies operate selectively by infecting target groups of neurons with a non-replicating retro-virus that carries a particular gene — a channelrhodopsin in the case of optogenetics. This gene will be integrated into the genome of the infected neurons and expressed, thereby introducing mutations with spatial specificity. In the case of optogenetics, the result is the modification of an existing piece of molecular machinery — an ion channel that adjusts ion concentrations thereby controlling the initiation of action potentials — which can now be experimentally controlled by shining a light on the cell.
Light-controlled sensing, excitation and silencing of individual neurons represents a major advance in experimental neuroscience, however it still doesn’t address the problem of scale. And new technology for robotic placement of 3-D multi-cell probes and automated patch-clamp electrophysiology [87], while they offer incremental improvements in recording technology, won’t scale either. Might there be some way we could automate the collection of neural-state information to canvass much larger collections of neurons? A scalable alternative to other methods for sensing the state of large populations of neurons — methods that are either invasive or outright destructive to the neural tissue or have limited coverage or poor temporal or spatial resolution — will require the means of collecting data from millions if not billions of neurons simultaneously.
One might imagine using the existing network of neurons as a packet-switching communication network using some variant of TCP/IP to transmit small packets of information collected from neurons buried deep in the brain to more accessible regions on the brain’s surface or outside the CNS entirely. Eventually it might be possible to use some form of low-power visible-light or radio-frequency transmission to transfer information through the dura and skull. However the technology to build the requisite nanoscale devices that both sense and transmit data in such a fashion is currently well beyond the state of the art. Alternatively and perhaps more practical in the near-term, we might use the network of arteries, vessels and capillaries that supply blood to the brain as a transportation and communication network to deliver much simpler nanoscale information-gathering machines to individual neurons and subsequently collect either the machines themselves or their encapsulated-information products to read off the state of neurons.
The blood supply to the brain is protected by a blood brain barrier which helps to avoid toxins and contaminants from damaging the brain. It does so by restricting the “diffusion of microscopic objects [e.g., bacteria], and large or hydrophilic molecules into the cerebrospinal fluid, while allowing the diffusion of small hydrophobic molecules [e.g., O2, CO2 and hormones such as melatonin]. Cells of the barrier actively transport metabolic products such as glucose across the barrier with specific proteins.” To accomplish this feat, the lining of the capillaries supplying blood to the brain consist of “high-density cells restricting passage of substances from the bloodstream much more than endothelial cells in capillaries elsewhere in the body.” While this action is generally beneficial to the organism, it can complicate the design of drugs and other interventions that directly target cortical tissue.
The interplay between neurons and the blood supply is mediated by a class of glial cells called astrocytes which perform many functions, including “biochemical support of endothelial cells that form the blood-brain barrier, provision of nutrients to the nervous tissue, maintenance of extracellular ion balance, and a role in the repair and scarring process of the brain and spinal cord following traumatic injuries.” We propose to employ a method of cellular subterfuge by disguising nanoscale machine payloads as normal nutrient packets to pass through endothelial gauntlet and then coopt the transport machinery of the astrocytes to deliver the packages to the intended target neurons. Packets of information disguised as waste products will pass back across the neural membranes into the cytoplasm, there to be scavenged by the lymphatic system and returned into circulation in the blood supply.
These information packets once they are released back into into the blood stream would be intercepted by a cell sorter located in the periphery and shunted through a processor that would read out the encoded information and convey it to a device outside the organism using some form of light or radio transmission. The packet processor and transmission machinery needn’t conform to the same size and immune-rejection requirements as the instruction packets that must pass through the blood brain barrier. Once collected and collated, the neural-state information can be processed to support a variety of genomic, connectomic and proteomic analyses.
In his lecture entitled “There’s Plenty of Room at the Bottom”, Richard Feynman considered the idea of “swallowing the doctor” which he credited his friend and graduate student Albert Hibbs. “This concept involved building a tiny, swallowable surgical robot by developing a set of one-quarter-scale manipulator hands slaved to the operator’s hands to build one-quarter scale machine tools analogous to those found in any machine shop. This set of small tools would then be used by the small hands to build and operate ten sets of one-sixteenth-scale hands and tools, and so forth, culminating in perhaps a billion tiny factories to achieve massively parallel operations. He uses the analogy of a pantograph as a way of scaling down items.” — see also Feynman’s tiny machines lecture.90 Eric Drexler’s PhD thesis at MIT has a good discussion of scaling laws relevant to classical dynamics and how the forces of consequence change as we approach realm of nanoscale machines.
Synthetic biologists are getting better at designing circuits using genes. We’ll see how quickly they are able to advance the state of the art to the point where they can create complex circuits that challenge existing silicon-based circuits while demonstrating the potential of biological compatability. In the meantime, cell bodies are full of molecular machinery that we can use directly or adapt for our particular purposes. For the proposal being considered here, we would need molecular sensors to detect what proteins are being transcribed and editing tools to splice information about what, when and where the proteins are being produced into cell products that would be reintroduced into the bloodstream to be carried out of the brain where they can be read out by larger molecular machines that would be more difficult to slip through the blood brain barrier.
To reconstruct a complete picture of what’s going on in the brain, we would need to encode information about which cells are producing what proteins and when. It may be that we can generate individual retroviruses each with a unique identifier — a promoter-tagged pointer to a random fixed-length sequence of DNA that would serve as a signature for a given cell, though it wouldn’t surprise me if randomness is as hard to come by in biological systems as it is in modern computing practice. Fortunately, some reasonable approximation to random will serve fine, since we anticipate the bulk of the data processing will be carried out externally where we can bring statistical algorithms to bear in sorting out identifiers that map to more than one cell. Getting accurate timing information could be tricky. However, there are rhythmic neural signals — beta: 13–30 Hz; alpha 8–13 Hz; theta 4–7 Hz; delta 0.5 to 4 Hz — that could possibly be used in combination to synchronize cellular clocks to within a few milliseconds.
November 16, 2012
I’ve given this log short shrift during the quarter in large part because I’ve been dedicating my time and effort and, in particular, my prose hacking to our CVPR paper on Drishti which was submitted yesterday. In this log entry, I’m going to include some notes that I’ve circulated in various venues, in an effort to consolidate them in a single place for subsequent reference.
A new gene has been discovered and claimed to be responsible for key differences between apes and humans. The authors of the study [70] claim that the gene is “highly active in two areas of the brain that control our decision making and language abilities. The study suggests it could have a role in the advanced brain functions that make us human.” This paper is interesting for how data in the form of sequences from different species, different populations and even from extinct lineages plus the ability quickly obtain new sequences, manipulate expression and transcription experimentally have made this sort of study possible. The discussion combines detailed information on biological processes along with arcana relating to current technology for obtaining such data.
Feynman’s lectures on tiny machines start with examining the challenge of writing the text of all the books in the Caltech library on the head of a pin, describes how conventional light-based lithography is used in the manufacture of computer chips — see also this Intel video, proceeds to describe how the limitations of this method pertaining to the wavelength of light can be overcome using other methods, and then turns to his central topic of making tiny machines which today is part of the growing field of nanotechnology and in particular the sub field of micromechanical systems or MEMS.92 At some point, Feynman asks the audience how he knows we can build these tiny machines. After a facetious remark about his being a physicist and knowing the laws of physics he checked and it’s possible, he said that a better answer is that “Living things have already done it. Bacteria swim through water at a scale it would be like us swimming through thick goop.” By the way, only first half of the video of the “Tiny Machines” lecture is the actual lecture; in the remaining half he answers questions many of which are pretty wide of the mark, but if you persist, you’ll find some gems in his answers to the more reasonable questions. If you’d like to learn more about nanotechnology, you might check out Stanford professor Kathryn Moler’s lectures.
Winfried Denk and his colleagues at the Max Planck Institute have developed a new tissue preparation and staining method that allows them to prepare large pieces of tissue for tracing out circuits in the mouse brain. The traditional approach of sectioning the tissue into small blocks, scanning them individually and then fitting the pieces back together is tedious and error prone, even when using automated methods for tracing out axons, most of which are “less than one micron thick, some even smaller than 100 nanometers.” Their method of block-face microscopy overcomes this problem by inserting an entire piece of tissue in the microscope and scanning the surface. Only then is a thin section cut, and the layer below is scanned. This makes it easier to combine the data on the computer.” It’s worth contrasting Denk’s method with the one being pursued by Sebastian Seung and his team at MIT. A summary of the article [110] in Nature Methods is available here.
Finally here are some pointers to questions, new research results and interesting references related to biological computing and computational neuroscience that caught my eye as possible fodder for a talk I’m giving at Stanford right after the Thanksgiving break:
How do biological circuits sense their environment, e.g., signals from other cells, signals from “distant” locations within the same cell, “foreign” material whether that be proteins, toxins, DNA from bacteria and viruses?
How do cells manufacture products required for their proper function centrally, e.g., inside the nucleus, the endoplastic reticulum or other organelles, transport manufactured products, e.g., using microtubules, or arrange for their remote production or assembly in distributed locations throughout the cell?
How do neurons perform computations that don’t directly involve the central cell body or soma, e.g., in circuits comprised entirely of dendrites — dendro-dendritic processing?
New method of decoding neural circuit designs from examining calcium fluorescence measurements.93
MIT team builds the most complex synthetic-biology circuit yet.
Petreanu et al [125] describe experiments on mice in which they are able to map long-range callosal projections using techniques from the optogenetic toolkit.
Researchers claim to have discovered key molecules involved in forming long-term memories — also featured in a Science Friday interview.
Interesting example demonstrating how a better algorithm [89] was able to substantially improve the performance of 3rd generation nanopore sequencing [149].
Signalling in plants where one leaf sends out chemical signals that presumably diffuse throughout the plant causing other leaves to manufacture toxins to discourage further damage — Rahul mentioned this in the context of considering how light might be used to stimulate the brain.
Here is Bill Freeman’s webpage for his Eulerian Video Magnification work. You can find a few interesting demo video, Matlab code, and publication. MindFlex is a company who makes simple EEG sensors for games.
A consortium of EU researchers combined 3-D images from the MRI scans of 100 brains of volunteers to create an atlas describing the white-matter connections using a new analysis tools and better diffusion-tensor imaging technology.94 Presumably the subjects were of approximately the same age and preferably older than twenty five, given that there are dramatic differences in the white-matter distributions across a wide range of ages and, in particular, over the long developmental period in primates ranging from birth into middle or late twenties.
A RadioLab series on our fragmented minds and asking the question “Who am I?” — with pieces from V.S. Ramachandran on the evolution of human consciousness, Robert Sapolsky integrating memories of his father, Robert Lewis Stevenson’s unpublished stories about dreaming and Dr. Jekyll and Mr. Hyde, and Paul Brok, author of the Into the Silent Land, invitation into the world of his childhood inhabited by “the little people” he had no control over.
On listening to Caroll Barnes lecture on the aging brain I started thinking about the how the ratio of white to grey matter changes over time and what the informational and metabolic consequences are for what the brain can compute on a fixed energy budget. I was also inspired by one of her slides to re-consider connectomics using fluorescent proteins in brainbow as an alternative to traditional staining in identifying neurons and their processes; current state of the art allows over one hundred differently mapped neurons to be simultaneously and differentially illuminated in this manner.
November 9, 2012
In answer to a question about cache reference, Mark Segal pointed me to this list of numbers that every software engineer should know compiled by Jeff Dean:
L1 cache reference 0.5 ns
Branch mispredict 5 ns
L2 cache reference 7 ns
Mutex lock/unlock 100 ns
Main memory reference 100 ns
Compress 1K bytes with Zippy 10,000 ns
Send 2K bytes over 1 Gbps network 20,000 ns
Read 1 MB sequentially from memory 250,000 ns
Round trip within same datacenter 500,000 ns
Disk seek 10,000,000 ns
Read 1 MB sequentially from network 10,000,000 ns
Read 1 MB sequentially from disk 30,000,000 ns
Send packet from CA to Netherlands and back to CA 150,000,000 ns
While not nearly so useful here’s a page full of facts about brains courtesy of Eric Chudler at the University of Washington — he refers to his compilation as neural numeracy.
November 27, 2012
I expect that many of the students in Yoav’s class were bewildered by much of what I had to say. The material in my lecture covered several disciplines and some of the points I made are subtle and take some getting used to. I suggested that Yoav challenge the students interested in learning more to conduct the following exercise: Read the abstract of the 2005 paper on optogenetics by[23] and then go straight to the “Methods” section. Read each sentence in sequence and if there’s any term you don’t understand then look it up on Wikipedia; create an appendix in anticipation of writing a short precis summarizing Boyden’s paper and containing each term you had to look up along with an abbreviated definition in your own words.
In reading the Wikipedia entries, you will likely encounter additional terms you are unfamiliar and so recurse until you feel comfortable with your understanding of the term you started with. When you’re done you’ll have a tree — or perhaps forest — of relevant terms. Along the way, take the time to try to answer questions that come up such as “Why do they work with hippocampal cells?” — if you answer this particular question you’ll learn something about the life cycle of neurons. When your appendix is reasonably complete, go back to the abstract — or to the methods section if you’re more ambitious — and rewrite the abstract in your own words expanding concepts that you wish were made clearer when you first read it. I guarantee that if you carry out this exercise you’ll learn a great deal and you’ll retain that knowledge longer and in more detail than is possible with less focused learning.
I do this sort of focused reading all the time and have discovered that not only does it improve my understanding but I find it much less frustrating, since, rather than trying to grasp some complex idea all at once, I am constantly learning bits and pieces of the larger puzzle and integrating them into the rest of what I know about the subject. It can take a fair bit of time to read a paper in a discipline you’re unfamiliar with but the method quickly pays dividends and soon you’ve compiled a glossary and set of references that you can easily navigate in and you’ll be reading and understanding new papers much more quickly. A while back, I read selected chapters in “From Genes to Genomes: Concepts and Applications of DNA Technology” by Jeremy Dale, Malcolm von Schantz and Nick Plant, and here’s a partial listing of the items that I compiled in reading the first chapter:
A plasmid is “a small DNA molecule that is physically separate from, and can replicate independently of, chromosomal DNA within a cell. [...] Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms.”
A vector is “a DNA molecule used as a vehicle to transfer foreign genetic material into another cell. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Common to all engineered vectors are an origin of replication, a multicloning site, and a selectable marker.”
A selectable marker is “a gene introduced into a cell [...] that confers a trait suitable for artificial selection. They are a type of reporter gene used [...] to indicate the success of a [...] procedure meant to introduce foreign DNA into a cell [and include] antibiotic resistance genes.”
A ligase (DNA or RNA) is “is a specific type of enzyme, a ligase, that facilitates the joining of DNA [RNA] strands together by catalyzing the formation of a phosphodiester bond. [...] It plays a role in repairing single-strand breaks in duplex DNA in living organisms.”
A polymerase chain reaction (PCR) is “an extremely versatile technique for copying DNA. [...] PCR allows a single DNA sequence to be copied (millions of times), or altered in predetermined ways.”
A restriction enzyme (or restriction endonuclease) is “an enzyme that cuts DNA at specific recognition nucleotide sequences known as restriction sites. [...] Inside a bacteria, the restriction enzymes selectively cut up foreign DNA in a process called restriction; while host DNA is protected by a modification enzyme (a methylase) that modifies the bacterial DNA and blocks cleavage.”
A bacteriophage or just phage is “a virus that infects and replicates within bacteria. [...] Bacteriophage are composed of proteins that encapsulate a DNA or RNA genome [and] replicate within bacteria following the injection of their genome into the cytoplasm.”
Following transplantation into the host, the foreign “DNA contained within the recombinant DNA construct may or may not be expressed. DNA may simply be replicated without expression, or it may be transcribed and translated so that a recombinant protein is produced [which may require] restructuring the gene to include sequences that are required for producing a mRNA molecule that can be used by the host’s translational apparatus [promoter, translational initiation signal, and transcriptional terminator].”
A nanopore is “a small hole, of the order of 1 nanometer in internal diameter. Certain porous transmembrane cellular proteins act as nanopores, and nanopores have also been made by etching a somewhat larger hole (several tens of nanometers) in a piece of silicon, and then gradually filling it in using ion-beam sculpting methods which results in a much smaller diameter hole: the nanopore. Graphene is also being explored as a synthetic substrate for solid-state nanopores.”
November 25, 2012
Since first thinking about the idea a couple of days ago, I’ve come up with several pages of written notes, half a dozen alternative “algorithms” and a bunch of pointers to related ideas both speculative — from futurists and science fiction writers — and provocatively practical — from scientists and engineers. Several science fiction writers and futurists have considered the idea of delivering nanoscale machines to the brain via the blood supply, e.g., in a 2009 interview Ray Kurzweil suggested:
By the late 2020s, nanobots in our brain (that will get there noninvasively, through the capillaries) will create full-immersion virtual-reality environments from within the nervous system. So if you want to go into virtual reality the nanobots shut down the signals coming from your real senses and replace them with the signals that your brain would be receiving if you were actually in the virtual environment. So this will provide full-immersion virtual reality incorporating all of the senses.and I expect that Kurzweil, Hans Moravec and probably quite a few other futurists and science fiction writers have spun similar stories in print since the publication of Eric Drexler’s Engines of Creation in 1986. (See the article “The Singularity is Far: A Neuroscientist’s View” for a contrarian view written by a neuroscientist.) There is also quite a bit of buzz — techno babble really — about using some form of nanotechnology to knock out cancer cells, overcome drug addiction and cure warts — just kidding about the last. While perhaps a detail to reporters, the current proposal is seriously considering nanoscale machines designed from scratch but rather adapting existing molecular machinery in much the same way as optogenetics uses recombinant DNA technology to adapt voltage-gated ion channels to open or close in response to a light signal.
It may also be possible to tag molecules in such a way to record biological events that would be sufficient to infer interesting global properties of neuron ensembles if only we could transmit the information to a computer for processing. There are on the order of 1014 edges (synapses) in the human cortical connectome, on the order of 1011 nodes (neurons) and, say, a few hundred neurotransmitters responsible for most of what goes on in the brain. We can encode any integer between 0 and 18,446,744,073,709,551,615 in 64 bits. In principle, we could tag each neuron, each of its synapses, and each class of neurotransmitters with a unique 64 bit integer. What if could record every event (or a representative sample of such events) of the form: neurotransmitter X originating from neuron A was received at neuron B at time T, encode the information in an RNA sequence, encapsulate it in a protective protein shell like a viral capsid, and boot it out of the CNS appropriately tagged so it can be identified and filtered out of the blood stream for analysis.
On the topic of using sequencing to infer connectomic information, Zador et al [172] have recast the study of neural circuitry as a problem of high-throughput DNA sequencing. The authors claim that such a method has the potential to increase efficiency by orders of magnitude over methods that depend on light or electron microscopy. My concerns about passing retro-viruses through the blood brain barrier were not entirely unfounded, but nature has figured out workarounds to deliver some of its scarier brain pathogens. The papers I found on the role of astrocytes and the endothelium suggest we might learn some tricks from these pathogens that could simplify the sensing interface.95 Speaking of nasty pathogens, check out the Wikipedia page, YouTube video and New York Times article on viral-based neuronal circuit tracing — the virus in this case is rabies. Unlike the approach that we’ve been considering in our previous posts, this method uses a self-replicating viral vector to propagate markers throughout the cell and to adjacent cells connected through axonal processes.96
November 21, 2012
The Brainbow technology being developed at Harvard is an alternative or possibly a useful complement to the approach pursued by Sebastian Seung [136] for constructing the connectome of an organism’s brain or other neural tissues such as the retina. It is a “process by which individual neurons in the brain can be distinguished from neighboring neurons using fluorescent proteins. By randomly expressing different ratios of red, green, and blue derivatives of green fluorescent protein in individual neurons, it is possible to flag each neuron with a distinctive color. This process has been a major contribution to the field of connectomics, or the study of neural connections in the brain.” This approach has a number of drawbacks including the requirement of breeding at least two strains of transgenic animals from embryonic stem cells which is both time consuming and complex.
Optogenetics is “the integration of optics and genetics to control precisely defined events within specific cells of living tissue even within freely moving animals, with the temporal precision (millisecond-timescale) needed to keep pace with functioning intact biological systems.” I mention it in the context of reading brain states because the development of optogenetic technology necessarily includes the development of “genetic targeting strategies such as cell-specific promoters or other customized conditionally-active viruses, to deliver the light-sensitive probes to specific populations of neurons in the brain of living animals, e.g. worms, fruit flies, mice, rats, and monkeys.” These same developments would serve as technology enablers for the sort of non-invasive, neural-state sensing that I’m proposing — see Bernstein and Boyden [16] for a discussion of the optogenetic toolkit.
Specifically the channelrhodopsins used in optogenetics can be readily expressed in excitable cells such as neurons using a variety of transfection techniques, e.g., viral transduction, cell-plasma-membrane electroporation. In the case of optogenetics, the channelrhodopsins function as light-gated ion channels. The so-called C-terminus of the channelrhodopsin extends into the intracellular space and can be replaced by fluorescent proteins without affecting channel function. Researchers are already developing light-controlled probes using related technology that supports sensing brain states [101].
Optogenetic technologies operate selectively by infecting target groups of neurons with a non-replicating retro-virus that carries a particular gene — a channelrhodopsin in the case of optogenetics. This gene will be integrated into the genome of the infected neurons and expressed, thereby introducing mutations with spatial specificity. In the case of optogenetics, the result is the modification of an existing piece of molecular machinery — an ion channel that adjusts ion concentrations thereby controlling the initiation of action potentials — which can now be experimentally controlled by shining a light on the cell.
Light-controlled sensing, excitation and silencing of individual neurons represents a major advance in experimental neuroscience, however it still doesn’t address the problem of scale. And new technology for robotic placement of 3-D multi-cell probes and automated patch-clamp electrophysiology [87], while they offer incremental improvements in recording technology, won’t scale either. Might there be some way we could automate the collection of neural-state information to canvass much larger collections of neurons? A scalable alternative to other methods for sensing the state of large populations of neurons — methods that are either invasive or outright destructive to the neural tissue or have limited coverage or poor temporal or spatial resolution — will require the means of collecting data from millions if not billions of neurons simultaneously.
One might imagine using the existing network of neurons as a packet-switching communication network using some variant of TCP/IP to transmit small packets of information collected from neurons buried deep in the brain to more accessible regions on the brain’s surface or outside the CNS entirely. Eventually it might be possible to use some form of low-power visible-light or radio-frequency transmission to transfer information through the dura and skull. However the technology to build the requisite nanoscale devices that both sense and transmit data in such a fashion is currently well beyond the state of the art. Alternatively and perhaps more practical in the near-term, we might use the network of arteries, vessels and capillaries that supply blood to the brain as a transportation and communication network to deliver much simpler nanoscale information-gathering machines to individual neurons and subsequently collect either the machines themselves or their encapsulated-information products to read off the state of neurons.
The blood supply to the brain is protected by a blood brain barrier which helps to avoid toxins and contaminants from damaging the brain. It does so by restricting the “diffusion of microscopic objects [e.g., bacteria], and large or hydrophilic molecules into the cerebrospinal fluid, while allowing the diffusion of small hydrophobic molecules [e.g., O2, CO2 and hormones such as melatonin]. Cells of the barrier actively transport metabolic products such as glucose across the barrier with specific proteins.” To accomplish this feat, the lining of the capillaries supplying blood to the brain consist of “high-density cells restricting passage of substances from the bloodstream much more than endothelial cells in capillaries elsewhere in the body.” While this action is generally beneficial to the organism, it can complicate the design of drugs and other interventions that directly target cortical tissue.
The interplay between neurons and the blood supply is mediated by a class of glial cells called astrocytes which perform many functions, including “biochemical support of endothelial cells that form the blood-brain barrier, provision of nutrients to the nervous tissue, maintenance of extracellular ion balance, and a role in the repair and scarring process of the brain and spinal cord following traumatic injuries.” We propose to employ a method of cellular subterfuge by disguising nanoscale machine payloads as normal nutrient packets to pass through endothelial gauntlet and then coopt the transport machinery of the astrocytes to deliver the packages to the intended target neurons. Packets of information disguised as waste products will pass back across the neural membranes into the cytoplasm, there to be scavenged by the lymphatic system and returned into circulation in the blood supply.
These information packets once they are released back into into the blood stream would be intercepted by a cell sorter located in the periphery and shunted through a processor that would read out the encoded information and convey it to a device outside the organism using some form of light or radio transmission. The packet processor and transmission machinery needn’t conform to the same size and immune-rejection requirements as the instruction packets that must pass through the blood brain barrier. Once collected and collated, the neural-state information can be processed to support a variety of genomic, connectomic and proteomic analyses.
In his lecture entitled “There’s Plenty of Room at the Bottom”, Richard Feynman considered the idea of “swallowing the doctor” which he credited his friend and graduate student Albert Hibbs. “This concept involved building a tiny, swallowable surgical robot by developing a set of one-quarter-scale manipulator hands slaved to the operator’s hands to build one-quarter scale machine tools analogous to those found in any machine shop. This set of small tools would then be used by the small hands to build and operate ten sets of one-sixteenth-scale hands and tools, and so forth, culminating in perhaps a billion tiny factories to achieve massively parallel operations. He uses the analogy of a pantograph as a way of scaling down items.” — see also Feynman’s tiny machines lecture.97 Eric Drexler’s PhD thesis at MIT has a good discussion of scaling laws relevant to classical dynamics and how the forces of consequence change as we approach realm of nanoscale machines.
Synthetic biologists are getting better at designing circuits using genes. We’ll see how quickly they are able to advance the state of the art to the point where they can create complex circuits that challenge existing silicon-based circuits while demonstrating the potential of biological compatability. In the meantime, cell bodies are full of molecular machinery that we can use directly or adapt for our particular purposes using recombinant DNA technology. For the proposal being considered here, we would need molecular sensors to detect what proteins are being transcribed and editing tools to splice information about what, when and where the proteins are being produced into cell products that would be reintroduced into the bloodstream to be carried out of the brain where they can be read out by larger molecular machines that would be more difficult to slip through the blood brain barrier.
To reconstruct a complete picture of what’s going on in the brain, we would need to encode information about which cells are producing what proteins and when. It may be that we can generate individual retro-viruses each with a unique identifier — a promoter-tagged pointer to a random fixed-length sequence of DNA that would serve as a signature for a given cell, though it wouldn’t surprise me if randomness is as hard to come by in biological systems as it is in modern computing practice. Fortunately, some reasonable approximation to random will serve fine, since we anticipate the bulk of the data processing will be carried out externally where we can bring statistical algorithms to bear in sorting out identifiers that map to more than one cell. Getting accurate timing information could be tricky. However, there are rhythmic neural signals — beta: 13–30 Hz; alpha 8–13 Hz; theta 4–7 Hz; delta 0.5 to 4 Hz — that could possibly be used in combination to synchronize cellular clocks to within a few milliseconds.
Selected quantitative facts about the human brain from Eric Chudler’s compilation:
Average number of neurons in the brain = 100 billion (1011)
Ratio of grey to white matter = [1.3, 1.1, 1.5] by age [20, 50, 100]99
Percentage of cerebral oxygen consumption by white matter = 6%
Percentage of cerebral oxygen consumption by gray matter = 94%
Number of neocortical neurons 20 billion (1010)100
Average loss of neocortical neurons = 100,000 per day (105)101
Number of synapses in cortex = 0.1 quadrillion (1014)
Total surface area of the cerebral cortex = 2,500 cm2
Percentage of cortical volume: frontal = 41%, temporal = 22%; parietal = 19%; occipital = 18%
Number of cortical layers = 6
Thickness of cerebral cortex = 1.5–4.5 mm
Number of fibers on the corpus callosum 250,000,000 (108)
Surface area of cerebellar cortex = 50,000 cm2
Number of fibers in human optic nerve = 1,000,000 (106)
Number of synapses for a “typical” neuron = 1,000 to 10,000
Diameter of neuron = 4-100 μm2 [granule/motor]
Single sodium pump maximum transport rate = 200 Na ions/sec; 130 K ions/sec
Typical number of sodium pumps = 1000 pumps per μm2 of membrane
Total number of sodium pumps for a small neuron = 1 million
Number of voltage-gated sodium channels in unmyelinated axon = 100 to 200 per μm2
Number of voltage-gated sodium channels at each node = 1,000 to 2,000 per μm2
Number of voltage-gated sodium channels between nodes = 25 per μm2
Membrane surface area of a “typical” neuron = 250,000 μm2
Membrane surface area of 100 billion neurons = 25,000 m2 (four soccer fields)
Typical synaptic cleft distance = 20-40 nanometers across
Number of molecules of neurotransmitter in one synaptic vesicle = 5,000
Action potential conduction rate [1, 10, 100] m/sec by diameter [0.1, 1, 10] μm102
Thinking about conventional computing hardware from etched traces and gates on silicon dies to servers and data centers:
Simple computing elements gates (AND, OR, NOT, NAND), more complicated (MULT, SHIFT, ALU, SIMD LANE).
Simple informational units (BIT, BYTE, CHAR, SINGLE, DOUBLE), more complicated (STRUCT, ARRAY, LIST).
Simple memory units (REGISTER, RAM, L1/L2 CACHE), more complicated (LOCAL DISK, RAID, MAGNETIC TAPE).
Simple communication units (WIRE, TRACE, BUS), more complicated (UART, ATA, SCSI, NIC).
How powerful are the units, how fast do they compute, how accurate are they and do they require additional processing?
How reliable are the units, how do they operate as they age, are they consistent, does temperature affect their output?
How much energy / material do they require, neurons need various molecules to manufacture proteins, neurotransmitters?
How much heat do they dissipate when idle and working and are there trade-offs relative to temperature and reliability?
How is information conveyed from one computing / memory unit to another, energy, reliability, locality, transfer speed?
How many units can you place in a cubic millimeter of space assuming different unit-to-unit communication trade-offs?
It is a useful exercise to think about the semiconductor fabrication processes that make these abstractions practical. The size of a transistor in a server-grade processor circa 2008 was 45 nm, which is measured in terms of the line width for the conduction channel of a MOSFET. The area on the die taken up by a typical processor MOSFET transistor in 2008 was more like 60 nm × 80 nm = 4.8 μm2 including a portion of the isolation wells. The depth is about 20 nm.
Feynman’s There’s Plenty of Room at the Bottom lecture enjoined his audience to entertain thought experiments like, “Is it possible to write the Encyclopedia of Britannica or the text of all the books in the Library of Congress on the head of a pin.” On a more practical note, he provided a theoretical perspective on such questions as “How many molecules would be necessary to build a NAND gate?” and “What’s the minimum width of a wire in atoms that would be required to reliably convey a certain voltage or alternative representation for one bit of information.”
As you halve the size and minimum distance separating computing units on a silicon die you increase the density of your computing hardware quadratically; if in addition you can manufacture three-dimensional chips you can increase the density cubically; of course, from a practical standpoint, you have to deal with heat dissipation, power distribution, communication on the chip as well as getting information on and off the chip.
The further you have to convey information generally the more power it takes, the more heat it produces and longer the transmission time; as a general of strategy, you want to localize the information required for computation but such compute-bound computations are relatively rare. Engineers have been able to create a single-atom transistor, but it will be considerably more challenging to pack a bunch of single-atom transistors in a small space and wire them together to perform useful work. So called super-computing centers like Los Alamos National Labs often emphasize computations involving lots of floating point computations such as those required for solving the systems of equations required for simulating the physics associated with nuclear detonations. However, most of the energy spent in computation worldwide is involved in moving information around and the computation are dominated by logical binary and integer operations. What is the right level of abstraction for thinking about computation? The transistor, logic gate, CPU core, processor, circuit board, server, rack, network, data center. Let’s try to think how Google engineers think about scalable computation and information transport. We’ll do so by considering a biologically-inspired approach to computer vision that was featured in an article on Google Brain in the New York Times. But first let’s see if we can gain some insight into how industrial software engineers think about scaling computation.
Here is list of facts about the current generation of computing hardware that every software engineer should know. It was compiled by Jeff Dean at Google:
L1 cache reference 0.5 ns
Branch mispredict 5 ns
L2 cache reference 7 ns
Mutex lock/unlock 100 ns
Main memory reference 100 ns
Compress 1K bytes with Zippy 10,000 ns
Send 2K bytes over 1 Gbps network 20,000 ns
Read 1 MB sequentially from memory 250,000 ns
Round trip within same datacenter 500,000 ns
Disk seek 10,000,000 ns
Read 1 MB sequentially from network 10,000,000 ns
Read 1 MB sequentially from disk 30,000,000 ns
Send packet from CA to Netherlands and back to CA 150,000,000 ns
November 19, 2012
As an example of how scaling methods in chip fabrication have enabled scaling in genome sequencing, we’ll be talking about a bunch of new technologies many of which are featured in that are featured in Kevin Davies book entitled “The $1000 Genome”. There is also a nice summary of current technology in Eric Topol’s “Creative Destruction of Medicine”. If you want to learn more and don’t have access to either of these books, this video provides a basic introduction to gene sequencing and the Sanger method — the basic method for first generation sequencing technology.
The human genome is about 3 billion base pairs. Due to the overhead of reassembling the genome from the pieces resulting from short read lengths — this is partly a limitation of the current biologically-based nucleotide read methods and partly a feature of the so-called shotgun method — sequencing the entire human genome requires approximately 13 coverage to achieve high accuracy. Perfect accuracy may not be possible due to the high frequency of repeated duplicate subsequences. The total number of base pairs that have to be read is on the order of 30 billion.
Second generation sequencing is best exemplified by a technique called pyrosequencing which largely replaced the Sanger method. The Illumina Genome Analyzer (Solexa) uses pyrosequencing and achieves 10 giga base pairs per run where each run requires 5 days with an average read length of 30 base pairs and a cost of $3K. The 454 (Roche) device manages 1 giga base pairs per run where each run requires 5 hours with an average read length of 500 base pairs and a cost of $5K.
Third generation sequencing hardware employs diverse methods but can be characterized as replacing much of the biology and chemistry with operations that can be performed in silicon. The Pacific Biosciences device uses a highly parallel method called single-molecule real-time sequencing, and Oxford Nanopore — which we’ll be talking about in class — takes the move to silicon one step further using a chip that reads electrical charge to determine each nucleotide as the DNA molecule threads a specially designed nanoscale pore.
November 17, 2012
Last year for Yoav’s class I had the students watch a video lecture entitled “Three Controversial Hypotheses Regarding Primate Brains” which eventually became the first lecture for the class that I offer in the Spring. One of the hypotheses that I consider in this lecture is due — at least in it’s most recent incarnation — to Robert Sapolsky from whom I understand you’ve already heard in this class. A somewhat more convenient HD version for those of you with high-speed internet makes it easy to select and search the slides and is available here.
This year I want to consider several related issues involving neuroscience and computer science, specifically concerning how computational scaling might accelerate our understanding of the human brain. By “computational scaling” I mean the science and technology involved in harnessing computation to achieve rapid, often exponentially increasing performance at a fixed or falling price point.
There is a good deal of hype surrounding computational scaling and it is often confused with other quickly accelerating technology trends like Gordon Moore’s Doubling Law for Transistor Density on VLSI chips — also known as “Moore’s Law” — and has been applied without adequate justification to everything from energy generation to food production. Ray Kurzweil has his variant called the Law of Accelerating Returns and Vernor Vinge’s Technological Singularity is an example of exponential process he predicts will undergo an phase transition or inflection point giving rise to a form of intelligence so far beyond humans that there will be no further basis for discourse between us and them.
Such speculation can make for fascinating or at least entertaining reading but they are not to be the subject matter of my lecture. Rather I want to investigate how we might go about understanding biological computation. It may be my use of the word “understand” does not fit with your definition and so I suggest that whenever I say “understand” you should feel free to substitute the phrase “simulate on Turing machine”.
I’ll start by describing a couple of projects my team at Google has been involved with and that were loosely inspired by biological models. In the process we’ll consider whether it would be possible to simulate a brain using the resources of an industrial data center like those maintained by Amazon, Google, Microsoft and other big players.
November 15, 2012
I’ve given this log short shrift during this quarter in large part because I’ve been dedicating my time and effort and, in particular, my prose hacking to our CVPR paper on Drishti which was submitted yesterday. In this log entry, I’m going to include some notes that I’ve circulated in various venues in an effort to consolidate them in a single place for subsequent reference.
A new gene has been discovered and claimed to be responsible for key differences between apes and humans. The authors of the study [70] claim that the gene is “highly active in two areas of the brain that control our decision making and language abilities. The study suggests it could have a role in the advanced brain functions that make us human.” This paper is interesting for how data in the form of sequences from different species, different populations and even from extinct lineages plus the ability quickly obtain new sequences, manipulate expression and transcription experimentally have made this sort of study possible. The discussion combines detailed information on biological processes along with arcana relating to current technology for obtaining such data.
Feynman’s lectures on tiny machines starts with writing the text of all the books in the Caltech library on the head of a pin, describes how conventional light-based lithography is used in the manufacture of computer chips — see also this Intel video, proceeds to describe how the limitations of this method pertaining to the wavelength of light can be overcome using other methods, and then turns to his central topic of making tiny machines which today is part of the growing field of nanotechnology and in particular the sub field of micro-mechanical systems or MEMS.103 At some point, Feynman asks the audience how he knows we can build these tiny machines. After a facetious remark about his being a physicist and knowing the laws of physics he checked and it’s possible, he said that a better answer is that “Living things have already done it. Bacteria swim through water at a scale it would be like us swimming through thick goop.” By the way, only first half of the video of the “Tiny Machines” lecture is the actual lecture; in the remaining half he answers questions many of which are pretty wide of the mark, but if you persist, you’ll find some gems in his answers to the more reasonable questions. If you’d like to learn more about nanotechnology, you might check out Stanford professor Kathryn Moler’s lectures.
On listening to Caroll Barnes lecture on the aging brain I started thinking about the how the ratio of white to grey matter changes over time and what the informational and metabolic consequences are for what the brain can compute on a fixed energy budget. I was also inspired by one of her slides to re-consider connectomics using fluorescent proteins in brainbow as an alternative to traditional staining in identifying neurons and their processes; current state of the art allows over one hundred differently mapped neurons to be simultaneously and differentially illuminated in this manner.
A RadioLab series on our fragmented minds and asking the question “Who am I?” — with pieces from V.S. Ramachandran on the evolution of human consciousness, Robert Sapolsky integrating memories of his father, Robert Lewis Stevenson’s unpublished stories about dreaming and Dr. Jekyll and Mr. Hyde, and Paul Brok, author of the Into the Silent Land, invitation into the world of his childhood inhabited by “the little people” he had no control over.
A consortium of EU researchers combined 3-D images from the MRI scans of 100 brains of volunteers to create an atlas describing the white-matter connections using a new analysis tools and better diffusion-tensor imaging technology.104 Presumably the subjects were of approximately the same age and preferably older than twenty five, given that there are dramatic differences in the white-matter distributions across a wide range of ages and, in particular, over the long developmental period in primates ranging from birth into middle or late twenties.
Winfried Denk and his colleagues at the Max Planck Institute have developed a new tissue preparation and staining method that allows them to prepare large pieces of tissue for tracing out circuits in the mouse brain. The traditional approach of sectioning the tissue into small blocks, scanning them individually and then fitting the pieces back together is tedious and error prone, even when using automated methods for tracing out axons, most of which are “less than one micron thick, some even smaller than 100 nanometers.” Their method of block-face microscopy overcomes this problem by inserting an entire piece of tissue in the microscope and scanning the surface. Only then is a thin section cut, and the layer below is scanned. This makes it easier to combine the data on the computer.” It’s worth contrasting Denk’s method with the one being pursued by Sebastian Seung and his team at MIT. A summary of the article [110] in Nature Methods is available here.
How do biological circuits sense their environment, e.g., signals from other cells, signals from “distant” locations within the same cell, “foreign” material whether that be proteins, toxins, DNA from bacteria and viruses?
How do cells manufacture products required for their proper function centrally, e.g., inside the nucleus, the endoplastic reticulum or other organelles, transport manufactured products, e.g., using microtubules, or arrange for their remote production or assembly in distributed locations throughout the cell?
How do neurons perform computations that don’t directly involve the central cell body or soma, e.g., in circuits comprised entirely of dendrites — dendro-dendritic processing?
New method of decoding circuit designs from calcium fluorescence measurements.105
MIT team builds the most complex synthetic-biology circuit yet.
Researchers claim to have discovered key molecules involved in forming long-term memories — also featured in a Science Friday interview.
Interesting example demonstrating how a better algorithm [89] was able to substantially improve the performance of 3rd generation nanopore sequencing [149].
Ray Kurzweil’s talk at Google plugging his book entitled How to Create a Mind: The Secret of Human Thought Revealed.
Signalling in plants where one leaf sends out chemical signals that presumably diffuse throughout the plant causing other leaves to manufacture toxins to discourage further damage — Rahul mentioned this in the context of considering how light might be used to stimulate the brain.
Here is Bill Freeman’s webpage for his Eulerian Video Magnification work. You can find a few interesting demo video, Matlab code, and publication. MindFlex is a company who makes simple EEG sensors for games.
Recasting the study of neural circuitry as a problem of high-throughput DNA sequencing instead of microscopy holds the potential to increase efficiency by orders of magnitude — Zador et al [172].
Speaker: David Bock, Affiliation: Lab Head, Janelia Farm Research Campus Howard Hughes Medical Institute, Host: Steven Smith, Title: “Neuronal network anatomy from large-scale electron microscopy”
The Economist The human microbiome: Me, myself, us
The microbiome, made much easier to study by new DNA-sequencing technology (which lets you distinguish between bugs without having to grow them on Petri dishes), is thus a trendy area of science. That, in itself, brings risks. It is possible that long-term neglect of the microbes within is being replaced by excessive respect, and that some of the medical importance now being imputed to the microbiome may prove misplaced. Whether or not that is true, though, there is no doubt that the microbiome does feed people, does help keep their metabolisms ticking over correctly and has at least some, and maybe many, ways of causing harm. And it may do one other thing: it may link the generations in previously unsuspected ways.A lot of the medical conditions the microbiome is being implicated in are puzzling. They seem to run in families, but no one can track down the genes involved. This may be because the effects are subtly spread between many different genes. But it may also be that some — maybe a fair few — of those genes are not to be found in the human genome at all. Though less reliably so than the genes in egg and sperm, microbiomes, too, can be inherited. Many bugs are picked up directly from the mother at birth. Others arrive shortly afterwards from the immediate environment. It is possible, therefore, that apparently genetic diseases whose causative genes cannot be located really are heritable, but that the genes which cause them are bacterial.
This is of more than merely intellectual interest. Known genetic diseases are often hard to treat and always incurable. The best that can be hoped for is a course of drugs for life. But the microbiome is medically accessible and manipulable in a way that the human genome is not. It can be modified, both with antibiotics and with transplants. If the microbiome does turn out to be as important as current research is hinting, then a whole new approach to treatment beckons.
The Economist Human genomics: The new world of DNA
When genes were first given a molecular basis, it was a fairly simple one. A gene was a piece of DNA that described a protein. When a cell had need of that protein it would cause a copy of the gene to be transcribed from DNA into RNA, a similar molecule capable of taking on more diverse forms. That RNA transcript would then be translated to make a protein. The bits of the genome which describe proteins this way have long been known to be only a fraction of the whole — a bit more than 1% — though it was accepted that some of the surrounding DNA was necessary to get the transcription machinery on and off the genes, thus turning them on or off as required. Human genes proved to be longer than might have been expected, with the RNA transcripts edited and rearranged before being translated into protein. Still, it seemed as if only a small fraction of the genome was actually doing anything, and that a lot of the rest was, or might as well be, “junk”.Now ENCODE has shown that fully three-quarters of the genome is transcribed into RNA at some stage in at least one of the body’s different types of cell. Some transcripts are whittled down more or less immediately, but 62% of the genome can end up in the form of a transcript that looks stable. There is a sense in which these transcripts are the basic constituents of the genome — its atoms, if you like. The transcripts which are associated with genes describing proteins are just one type among many.
All this RNA has a wide variety of uses. It regulates what genes actually make protein and how much is made in all sorts of complicated ways; some transcripts are millions of times more common than others. Even ENCODE has not been able to catalogue all of this diversity, but it has made headway in clarifying what to look for.
Whereas 62% of the genome may be turned into finished transcripts in some cell or other, only about 22% of the DNA ends up in such transcripts in the typical cell. This is because of molecular switches that turn parts of the genome on and off depending on what the cell in question is up to. Such switches are as worthy of their place in the parts list as the locations of particular regions that code for proteins. They are, though, harder to find — and, it turns out, much more numerous.
That you need a profusion of such switches to get the right pattern of genes turned on and off in a given cell at a given time is obvious. But the scale of the regulatory system has taken even some of its cartographers by surprise. Ewan Birney of the European Bioinformatics Institute, who was the lead coordinator of ENCODE’s data-analysis team, says he was shocked when he realised that the genome’s 20,000-odd protein-coding genes are controlled by some 4m switches.
Consumer BCI and EEG devices:
Zeo Sleep Manager is basically a three-channel EEG device. Here’s an interface that allows access to raw Zeo data and a useful forum discussing this library. Choudhury Ahmed attempts to get signals off the chip (source).
Pointers to two descriptor technologies that we might consider to the context work:
Serge Belongie’s shape-context [112]
Lazebnik et al spatial-pyramid matching [93, 170]
New finding on stress and stroke risk. The Sherpa Doctor Jay Parkinson. The Text to Matrix Matlab Toolbox. Theis et al [148, 147] on the limitations of deep belief networks.
Sunday, July 29, 2012
In the lecture entitled “Controversial Hypotheses in Computational Neuroscience”, I surveyed some of the primary sources of evidence for theories in computational neuroscience. In several of the cases mentioned, scientific theories get trotted out by the press and are judged by non-experts as was the case with Ernst Haeckle’s “Recapitulation Theory.” As an example of the sort of controversy that can erupt as consequence of such public airing, consider the case of Richard Dawkins critical review of Edward O. Wilson’s new book, “The Social Conquest of Earth”, in Prospect magazine made the news. In addition to his criticism of Wilson’s new popular-science book, Dawkins took the opportunity to trash the article [119] in Nature which Wilson cites as supporting the theory of group evolution presented in the book. Comment on this example and contrast with the publicity surrounding the discovery of the Higg’s boson — or at least something that behaves as we expect the Higg’s boson to behave.
When CS379C was taught in of Spring 2012, we looked at cutting-edge systems-neuroscience technologies being developed by Ed Boyden, Sebastian Seung and Steven Smith. CS379C in the Spring of 2013 will go back and check on their progress to see whether the assumed opportunities for scaling these technologies have been realized. H. G. Wells prophetically described various uses of atomic energy in The World Set Free and even was able to fairly accurately predict the time it would take to develop these technologies. Ray Kurzweil is well known for his predictions of technological breakthroughs by plotting the exponential growth of computing and then extrapolating to the year he thinks the computing power requisite for a given technology will be available or, in the case of consumer technology, affordable.
As a warm-up exercise on how to predict both the progress and prospects for the development of new technologies and the commercialization of existing ones, consider the issues concerning the following proposed and launched technology ideas: What about the commercial development and launching of geosynchronous satellites? It might make sense for Google, Microsoft or companies like MapQuest or Rand McNally that have to maintain up-to-date map data for driving directions. How would you analyze this potential? What about the proposed nuclear reactor technology called traveling wave reactor claimed to produce safe clean nuclear energy from depleted uranium, natural uranium, thorium or spent fuel removed from light water reactors?
How do engineers think about big projects and pushing existing technologies to unprecedented scale? At Google, Urs Hölzle is largely responsible for guiding the design and deployment of the technical infrastructure — hardware and related software — for Google’s globe-spanning collection of data centers. What are the most important challenges facing Google as it scales its capacity to serve users and offer new products to businesses requiring cloud-computing services. Consider an engineer’s perspective on Instant and Personalized Search. Comment on their potential as seen from an engineering perspective and the consequences as the technology is seen through the lens of the popular press which spins it to snare readership and reacts to and substantially influences popular opinion by so doing.
Rudyard Kipling wrote a series of so-called just-so stories for little children in which he provides entertaining but implausible accounts of how various phenomena came about. Perhaps your parents read to you from “How the Camel Got His Hump” or “How the Leopard Got his Spots” — you can read these and other stories at Project Gutenberg here. Kipling’s stories for children were not meant to be taken seriously, but idea of just-so-stories reveals a deeper need in humans to explain the world around them using simple — and often simplistic — causal stories that sound good but distort the truth and ignore the evidence. Daniel Kahneman describes this bias in his recent book [79] and as an example discusses the plausibility and accuracy of popular-press versions of the Google success story.
References
1 There are several mathematical models of spiking neurons designed to explain and predict the observed behavior of neurons — see Koch and Segev [86] or Dayan and Abbott [40] for good introductions. These include compartmental models of the sort that Henry Markram and his team at EPFL have developed for modeling cortical columns, as well as significant departures from the widely-used Hodgkin-Huxley model of signal propagation such as the soliton model which hypothesizes signals travel along the cell’s membrane in the form of acoustic pulses known as solitons. Source: Wikipedia
Many of the models involve application of cable theory in which dendrites and axons are modeled as cylinders composed of segments represented as clectrical circuits with capacitances cm and resistances rm combined in parallel:
 |
Electromagnetic theory is usually expressed using the standard International System of derived units (SI derived units). For the force F we use the newton, for the quantity of charge q we use the coulomb, the distances r are measured in meters, and time s is measured in seconds. The SI derived units for the electric field are volts per meter, newtons per coulomb, and teslas meters per second. Here is a brief review of the primary SI derived units relevant in understanding basic electrophysiology:
N — a newton is the SI measure for force equivalently 1 N = 1 kg × m/s2;
C — a coulomb is the SI measure of charge 6.24 × 1018 electrons (negative) protons (positive) equivalently 1 C = 1 A × 1 s;
V — a volt is the SI measure for electrical potential or voltage defined as the difference in electric potential across a wire when an electric current of one ampere dissipates one watt of power;
J — a joule is the SI measure for energy = 0.000000278 kilowatt-hours (kWh) = 0.287 milliwatt-hours (mWh) equivalently 1 J = N m = C V;
A — an ampere is the SI measure for electric current the amount of charge passing a point in a circuit per unit time with one coulomb per second constituting one ampere.
Ω — an ohm is the SI measure for resistance 1 Ω = 1 V / A — for a fixed voltage, the greater the resistance the smaller the current;
The following laws are used in analyzing the electrical circuits which are used to represent neurons and analyze how their intracellular and extracellular potentials change over time:
Coulomb’s Law | F | = k | q1 q2 | / r2 where q1 and q2 are measured in coulombs, | x | is the magnitude of x, and the constant k is dependent on the medium in which the charges are immersed. In the case of air, k is approximately 9.0 × 109 N m2 / C2. If the charged objects are present in water, the value of k can be reduced by as much as a factor of 80;
Ohm’s Law states that the current through a conductor between two points is directly proportional to the potential difference across the two points. This assumes that we hold the resistance of the conductor constant. Introducing the constant of proportionality, the resistance, we have the familiar formula: I = V / R;
Kirchoff’s Circuit Laws state that (1) the sum of currents flowing into a node of an electric circuit equal the sum of currents flowing out of that node, and (2) the sum of the voltages around any closed loop in an electric circuit is equal to zero assuming no fluctuating magnetic fields linking the closed loop;
Voltage is electric potential energy per unit charge, measured in joules per coulomb ( = volts). It is often referred to as “electric potential”, which then must be distinguished from electric potential energy by noting that the “potential” is a “per-unit-charge” quantity. Like mechanical potential energy, the zero of potential can be chosen at any point, so the difference in voltage is the quantity which is physically meaningful. The difference in voltage measured when moving from point A to point B is equal to the work which would have to be done, per unit charge, against the electric field to move the charge from A to B. The voltage between the two ends of a path is the total energy required to move a small electric charge along that path, divided by the magnitude of the charge.
2 In addition to discussing the ambiguity concerning the “imaging broadly construed” category. Anjali also opined “I’m guessing that some people would not consider EEG to be an imaging modality, but would of course consider it to be a diagnostic technology.” Here are my responses to Anjali’s questions:
That’s a good way of putting it. The “imaging” section is indeed about technologies in which the recording and reporting devices are external to the tissue and preferably external to the organism. MRI and MEG are obvious examples even though they don’t use light as the illumination medium. The fact that all of these technologies allow for external power and computation is one of the main reasons for their utility and accelerated development as clinical diagnostic tools.Most of the “imaging” technologies also produce images or maps of varying resolution, albeit 3-D voxel-based images. One exception is EEG, however it’s more-invasive cousin ECoG does produce a pretty credible low-resolution map though at the expensive of being more invasive since it requires a craniotomy while not actually penetrating brain tissue except in some of the more radical variants.
ECoG is the reason that I ended up putting EEG in the imaging (broadly construed) section; EEG is the better known technology, I wanted to mention it since it would be important in the discussion of BCI in the investment / applications sections, but clearly it lacks the resolution of the other technologies. Putting it in the imaging section was a compromise in keeping things simple and relative short. NIRS also falls in this category.
The picture gets muddier when you throw in organic dyes and contrast agents as these require more invasive alterations to the tissue sample. They do use light and produce images / maps however, and so it made sense to include mention of SEM, multi-photon EM, confocal microscopes, etc. in this section.
Keep in mind that you don’t have to cover all the technologies mentioned in the main text or, in particular, those mentioned the imaging section. Your job is to pick the most promising ones and provide the rationale and back-of-the-envelope calculations required to support your conclusions in predicting their future impact on BAM/BRAIN. MEG, PET, EEG, ECoG are off the table as far as leading contenders. MRI is definitely on the table and ultrasound-based technologies are the dark horses, gaining ground on the FUS-for-ablation-based-surgery side of things but still lagging on the imaging side due to resolution.
3 The lone axon emerging from the base of the pyramidal soma often extends over even longer distances, typically measured in tens of centimeters (e.g. one side of the brain to the other). The axon also branches profusely, thus resulting in many centimeters of total length.
 |
4 Ionic radius, is the radius of an atom’s ion. Although neither atoms nor ions have sharp boundaries, they are sometimes treated as if they were hard spheres with radii such that the sum of ionic radii of the cation and anion gives the distance between the ions in a crystal lattice. Ionic radii are typically given in units of either picometers (pm) or Ångströms, with 1 Ångström = 100 pm.
5 Parallel acquisition techniques combine the signals of several coil elements in a phased array to reconstruct the image, the chief objective being either to improve the signal-to-noise ratio or to accelerate acquisition and reduce scan time. (source)
6 Sydney Brenner figured prominently in the history of molecular biology and was a life-long friend and colleague of Francis Crick, having obtained a job at Cambridge in the Cavendish Unit through Crick’s efforts to recruit him and eventually joining Crick when he moved to the Salk Institute in La Jolla:
Sydney Brenner, Senior Distinguished Fellow of the Crick-Jacobs Center, Salk Institute for Biological Sciences, is one of the past century’s leading pioneers in genetics and molecular biology. Most recently, Brenner has been studying vertebrate gene and genome evolution. His work in this area has resulted in new ways of analyzing gene sequences, which has developed a new understanding of the evolution of vertebrates.You can learn more about Brenner from his autobiography entitled “My Life in Science”, or, for a shorter introduction, the autobiographical material provided as part of his Nobel Prize awarded in 2002 — see source. I also recently ran across Brenner’s “Loose Ends” column which was featured for many years in the journal Current Biology and you can find archived on the journal’s web site here.Among his many notable discoveries, Brenner established the existence of messenger RNA and demonstrated how the order of amino acids in proteins is determined. He also conducted pioneering work with the roundworm, a model organism now widely used to study genetics. His research with Caenorhabditis elegans garnered insights into aging, nerve cell function and controlled cell death, or apoptosis.
Much of the early history of molecular biology, while starting at Cambridge with Crick and Watson in the Cavendish lab, moved to the United States with Crick and Sydney Brenner eventually ending up at the Salk Institute and Watson, Max Delbruck and George Gamow at Cold Spring Harbor. These are colorful characters at a time in science when there were many larger than life personalities, many of whom were physicists pumped on their success in the first half of the 20th century and looking for the next big challenge to tackle. The ostensibly-autobiographical, popular-science “kiss-and-tell” was arguably born in this period with the publication of James Watson’s The Double Helix.
Watson was notoriously outspoken about the personal lives of the people he worked with, to the point where he upset many of these people with his rather extensive and revealing autobiographical writings. Francis Crick was much more reticent to reveal juicy tidbits about his colleagues or opine about their personalities in print. If you’re interested in a relatively unbiased view of the history of molecular biology you might try The Eighth Day of Creation: Makers of the Revolution in Biology by Horace Freeland Judson.
7 We have’t as yet talked about transcriptomics in this class, but it is clearly a field of study important in understanding the foundations neural signalling:
The transcriptome is the set of all RNA molecules, including mRNA, rRNA, tRNA, and other non-coding RNA produced in one or a population of cells. It differs from the exome in that it includes only those RNA molecules found in a specified cell population, and usually includes the amount or concentration of each RNA molecule in addition to the molecular identities. Source: Wikipedia
8 Researchers in Mark Schnitzer’s lab at Stanford are developing technology allowing the brain volumes of ~100 alert flies to be imaged simultaneously by two-photon fluorescence microscopy. They offer the following advantages of their approach (source):
The ability to track neural dynamics across the brains of large numbers of normal flies and those with genetically induced neural circuit perturbations will transform our understanding of how neural circuits produce animal behavior;
The now prominent role of the fruit fly as a model system for the study of developmental disorders, neurodegenerative diseases, and addiction implies we will gain significant medical insights into devastating conditions;
Our technology will have important applications to drug screening, allowing the cellular effects of new compounds to be assessed rapidly in vivo;
The ability to perform high-throughput time-lapse imaging of cellular events during the maturation of fly embryos will greatly benefit developmental neurobiology. Applications of our technology will also be plentiful in other model organisms such as nematodes and zebrafish, impacting multiple areas of biomedicine.
9 Roberto P.J. Barretto and Schnitzer, Mark J. In Vivo Optical Microendoscopy for Imaging Cells Lying Deep within Live Tissue. Cold Spring Harbor Protocols 2012 (PDF)
10 In our class discussion, Briggman commented on how critical resolution in the z axis is to successful circuit tracing. Justin Kinney from Ed Boyden’s lab mentioned that Kristen Harris at the University of Texas Austin believes Winfried Denk and Sebastian Seung can’t resolve sufficient detail with their imaging resolution. She manages 2-3nm in the x-y plane and 50nm in the z plane, and uses an extracellular stain that she claims makes all the difference in accurate cell-body segmentation;
11 See Recent Advances in Magnetic Resonance Imaging (PDF).
12 The new line of multiple-beam FIB-SEM (Focused Ion Beam Scanning Electron Microscopy) microscopes from Carl Zeiss promise to speed scanning times by as much as sixty-fold.
13 Here are two examples of simulation tools capable of generating biologically plausible data — at least plausible with respect to our current understanding of the underlying biophysics. The second example is called MCell and is capable of generating some of the most accurate simulations available to date. Unlike the attempts at simulating a single cortical column conducted by the EPFL / IBM Blue Brain collaboration, we envision starting with an accurate model of cytoarchitecture obtained using electron microscopy coupled with proteomic signatures synapses obtained from array tomography:
Michael J. Byrne, M. Neal Waxham, and Yoshihisa Kubota. Cellular Dynamic Simulator: An Event Driven Molecular Simulation Environment for Cellular Physiology. Neuroinformatics. 2010 June; 8(2): 63-82. (URL)
Rex A. Kerr and Thomas M. Bartol and Boris Kaminsky and Markus Dittrich and Jen-Chien Jack Chang and Scott B. Baden and Terrence J. Sejnowski and Joel R. Stiles. Fast Monte Carlo Simulation Methods for Biological Reaction-Diffusion Systems in Solution and on Surfaces. SIAM Journal Scientific Computing. 2008 October 13; 30(6): 3126. (URL)
14 Massachusetts Institute of Technology, Media Laboratory; Cambridge, MA — 1997–2004, Ph.D. in Media Arts and Sciences, Fall 2004. Thesis: Microslots: Scalable Electromagnetic Instrumentation. Advisor: Neil Gershenfeld.
15 An NMR device consists of a magnet and a transceiver antenna or probe that is placed inside the magnet. The design of the antenna determines the resolution of the device. Here’s an example of a paper discussing antenna design for high-resolution brain imaging:
This study introduced a localized approach for magnetic resonance microscopy (MRM) of the rat brain. A single-loop radiofrequency (RF) receiver coil designed for micro-imaging was developed by using electromagnetic simulation software widely used in communication fields. With transmit-only and receive-only (TORO) configuration, receive-only surface coil can achieve higher signal-to-noise ratio (SNR) at localized brain region. Corpus callosum and hippocampus were landmarks to evaluate the capacity of the proposed coil. On a 3T MRI system, high-resolution MRI of the dissected rat brain was acquired with spatial resolution of 117 × 117 × 500 μm3. The achieved high local SNR and spatial resolution will provide valuable information for resolving the architecture of the rat brain. Source: Localized High-resolution MR Imaging of Rat Brain Architecture Using Micro-fabricated Receive-only RF Coil. 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Meng-Chi Hsieh (2007).
16 An RFID device typically consists of a tag providing the electronics and an antenna, the two or which may or may not be packaged together. Antenna size, transmission frequency, required power and effective distance are coupled. As the electronics shrink, engineers are constantly trying to improve the efficiency and reduce the size of antennas. The minimum length for an efficient antenna is a half wavelength. This is called the dipole antenna. However, a short dipole is a physically feasible dipole formed by two conductors with a total length L very small compared with the wavelength λ.
The part of the spectrum occupied by TV and radio has wavelengths on the order of meters. You can increase the frequency into the microwave, but even so we’re talking about a λ of a millimeter for a 300GHz signal. Nanoscale RFID antennas can’t be that large. Instead, RFID antennas are loop antennas, consisting of tiny coil of wire. The size of the antenna, the material it’s made of, and the number of loops all influence the frequency range it can detect. This device has a tag 3.2 × 3.2 × 0.7mm operates at 13.56MHz and has an effective range of 15mm given an output of 200mW and an antenna size of 35 × 54mm.
17 Although the exact mechanism by which an RV infection causes a lethal neurological disease are still not well understood, the most significant factor underlying the lethal outcome of an RV infection appears to be the neuronal dysfunction due to drastically inhibited synthesis of proteins required in maintaining neuronal functions. Source: [48].
18 Understanding the role of short- and long-range connections in cortex and thalamus provides a good example [123] of the sort of pathology-related inquiry that would be facilitated by BAM/BRAIN instrumentation:
Although altered short-range cortical circuit function is thought to be the primary cause of injury-induced epilepsies, the role of long-range connections to and from injured cortex to other brain regions in seizure maintenance has not been adequately studied. The cortex is intimately connected with thalamus, and the cortico-thalamo-cortical excitatory loop mediates network oscillations underlying epilepsies in man and in animal models. (source)Chow and Boyden [34] discuss several other possible optogenetic interventions including a promising treatment for retinitis pigmentosa.
19 This might be appropriate, for example, in the case of the Alan Jasanoff’s NMR work described in this article in the quarterly report of the McGovern Institute for Brain Research at MIT.
20 Note that we have to deal with scattering in both the illumination and reflection steps.
21 Note that photoacoustic imaging laser illumination is differentially absorbed by tissues that thermoelastically expand emitting wide-band ultrasound which is detected by ultrasonic transducers. In this case, the tissue serves as its own recorder.
22 Note that coding and transmission can be powered by local cellular energy sources, e.g., ATP, or externally, e.g., resonant inductive coupling or mechanical energy transmitted via ultrasound.
23 Note that signal transmission can be push or pull with the transfer signal mediated by cellular events, asynchronous regular fixed-intervals, or via some external polling device such as focused scan.
24 This use of the term “fiddly bits” comes from Douglas Adams’s The Hitchhiker’s Guide to the Galaxy in which Slartibartfast, the engineer who designed Earth said his favorite part was doing the fiddly bits around the fjords.
25 Immunoflorescence uses the specificity of antibodies to their antigen to target fluorescent dyes to specific biomolecule targets within a cell, and therefore allows visualisation of the distribution of the target molecule through the sample. Standard immunoflorescence techniques are limited to fixed (i.e., dead) cells when structures within the cell are to be visualized because antibodies cannot cross the cell membrane. An alternative approach is using recombinant proteins containing fluorescent protein domains, e.g., green fluorescent protein (GFP). Use of such “tagged” proteins allows determination of their localization in live cells. Even though this seems to be an elegant alternative to immunofluorescence, the cells have to be transfected or transduced with the GFP-tag, and as a consequence they become at least S1 or above organisms that require stricter security standards in a laboratory. (source)
26 The fact that today’s microprocessor chips can have billions of transistors does considerably complicate matters.
27 In addition to cell type, e.g., in the visual cortex we find spiny pyramidal and stellate cells as well as a number of non-spiny neurons such as inhibitory basket cells, it is often useful in attempting to infer the function of a neural circuit (as Zador illustrates) to distinguish between inhibitory and excitatory neurons:
Active excitatory inputs result in excitatory postsynaptic potentials (EPSPs) through activation of AMPA and NMDA type glutamate receptors. Inhibitory inputs result in inhibitory postsynaptic potentials (IPSPs) through activation of GABAA or GABAB receptors. GABAA receptors can also mediate shunting inhibition.Excitatory and inhibitory inputs to pyramidal neurons are integrated in an ongoing manner to determine the output of the neuron, which is ultimately action potential firing in the axon. The integration of excitatory and inhibitory synaptic inputs is a complex process that depends on the magnitude and timing of the synaptic conductances as well as the spatial relationship between the activated synapses and the final integration zone in the axon. (source)
28 The ExAblate 4000 transcranial MRgFUS system (InSightec, Inc, Tirat Carmel, Israel) is a specialized system, integrating magnetic resonance imaging and high intensity focused ultrasound for investigational non-invasive, image-guided transcranial applications. The system enables intra-procedure MRI for therapy planning, and real-time MR thermal imaging feedback to monitor safety and efficacy. It is tightly integrated with an MR scanner and operates via a unique planning and treatment control workstation. MR imaging facilitates the accurate localization of the target region, safe delineation of the treatment margins, sustained real-time monitoring of tissue heating, and assessment of therapeutic outcome during and after therapy.
The ExAblate 4000 supports clinical studies with a unique patient interface, and provides tools for clinical, preclinical, in vivo and in vitro research. A hemispherical, helmet-like, multi-element phased array transducer enables focal targeting of brain tissue through the intact skull. The system is integrated with a standard GE MRI system using a detachable treatment table. In the scanner room, the patient lies on the table with their head immobilized in a stereotactic frame, and the helmet like transducer positioned around their head. A sealed water system with an active cooling and degassing capacity maintains the skull and skin surface at a comfortably low temperature.
The entire setup is moved into the MR scanner and a series of conventional MRI scans are displayed on the ExAblate workstation and analyzed by the attending physician to determine the targeted regions. Pre-operative CT and inter-operative MR scans are co-registered reconstructing a model of the skull and brain anatomy for treatment planning and simulation. The treatment is based on multiple sonications that cover the targeted volume. Sublethal spots confirm the target localization accuracy and patient comfort prior to lesion generation. During energy delivery to each spot, thermal images provide real-time feedback of the treatment location and measure thermal rise, allowing the physician to adjust the parameters accordingly. Post-treatment contrast imaging confirms the treatment effect. (Source)
29 PURPOSE: This study aims to demonstrate, using human cadavers the feasibility of energy-based adaptive focusing of ultrasonic waves using magnetic resonance acoustic radiation force imaging (MR-ARFI) in the framework of non-invasive transcranial high intensity focused ultrasound (HIFU) therapy.
METHODS: Energy-based adaptive focusing techniques were recently proposed in order to achieve aberration correction. The authors evaluate this method on a clinical brain HIFU system composed of 512 ultrasonic elements positioned inside a full body 1.5 T clinical magnetic resonance (MR) imaging system. Cadaver heads were mounted onto a clinical Leksell stereotactic frame. The ultrasonic wave intensity at the chosen location was indirectly estimated by the MR system measuring the local tissue displacement induced by the acoustic radiation force of the ultrasound (US) beams. For aberration correction, a set of spatially encoded ultrasonic waves was transmitted from the ultrasonic array and the resulting local displacements were estimated with the MR-ARFI sequence for each emitted beam. A noniterative inversion process was then performed in order to estimate the spatial phase aberrations induced by the cadaver skull. The procedure was first evaluated and optimized in a calf brain using a numerical aberrator mimicking human skull aberrations. The full method was then demonstrated using a fresh human cadaver head.
RESULTS: The corrected beam resulting from the direct inversion process was found to focus at the targeted location with an acoustic intensity 2.2 times higher than the conventional non corrected beam. In addition, this corrected beam was found to give an acoustic intensity 1.5 times higher than the focusing pattern obtained with an aberration correction using transcranial acoustic simulation-based on X-ray computed tomography (CT) scans.
CONCLUSIONS: The proposed technique achieved near optimal focusing in an intact human head for the first time. These findings confirm the strong potential of energy-based adaptive focusing of transcranial ultrasonic beams for clinical applications. (Source)
30 Unlike the spines, the surface of the soma is populated by voltage activated ion channels. These channels help transmit the signals generated by the dendrites. Emerging out from the soma is the axon hillock. This region is characterized by having an incredibly high concentration of voltage-activated sodium channels. In general, it is considered to be the spike initiation zone for action potentials (Source).
31 In contrast with an electrical synapse is a mechanical and electrically conductive link between two abutting neurons that is formed at a narrow gap between the pre- and postsynaptic neurons known as a gap junction. At gap junctions, such cells approach within about 3.5 nm of each other, a much shorter distance than the 20 to 40 nm distance that separates cells at a chemical synapse.
32 Genetically encoded calcium indicators (GECIs) are powerful tools for systems neuroscience. Recent efforts in protein engineering have significantly increased the performance of GECIs. The state-of-the art single-wavelength GECI, GCaMP3, has been deployed in a number of model organisms and can reliably detect three or more action potentials in short bursts in several systems in vivo. Through protein structure determination, targeted mutagenesis, high-throughput screening, and a battery of in vitro assays, we have increased the dynamic range of GCaMP3 by severalfold, creating a family of "GCaMP5" sensors. We tested GCaMP5s in several systems: cultured neurons and astrocytes, mouse retina, and in vivo in Caenorhabditis chemosensory neurons, Drosophila larval neuromuscular junction and adult antennal lobe, zebrafish retina and tectum, and mouse visual cortex. Signal-to-noise ratio was improved by at least 2- to 3-fold. In the visual cortex, two GCaMP5 variants detected twice as many visual stimulus-responsive cells as GCaMP3. By combining in vivo imaging with electrophysiology we show that GCaMP5 fluorescence provides a more reliable measure of neuronal activity than its predecessor GCaMP3. GCaMP5 allows more sensitive detection of neural activity in vivo and may find widespread applications for cellular imaging in general. Source: Akerboom J. and Chen T.W. and Wardill T.J. and Tian L. and Marvin J.S. and Mutlu S. and Calder�n N.C. and Esposti .F and Borghuis B.G. and Sun X.R. and Gordus A. and Orger M.B. and Portugues R. and Engert F. and Macklin J.J. and Filosa A. and Aggarwal A. and Kerr R.A. and Takagi R. and Kracun S. and Shigetomi E. and Khakh B.S. and Baier H. and Lagnado L. and Wang S.S. and Bargmann C.I. and Kimmel B.E. and Jayaraman V. and Svoboda K. and Kim D.S. and Schreiter E.R. and Looger L.L. (2012) Optimization of a GCaMP calcium indicator for neural activity imaging. The Journal of Neuroscience 32(40):13819-13840.
33 Two-photon calcium imaging of neuronal populations enables optical recording of spiking activity in living animals, but standard laser scanners are too slow to accurately determine spike times. Here we report in vivo imaging in mouse neocortex with greatly improved temporal resolution using random-access scanning with acousto-optic deflectors. We obtained fluorescence measurements from 34-91 layer 2/3 neurons at a 180-490 Hz sampling rate. We detected single action potential-evoked calcium transients with signal-to-noise ratios of 2-5 and determined spike times with near-millisecond precision and 5-15 ms confidence intervals. An automated ’peeling’ algorithm enabled reconstruction of complex spike trains from fluorescence traces up to 20-30 Hz frequency, uncovering spatiotemporal trial-to-trial variability of sensory responses in barrel cortex and visual cortex. By revealing spike sequences in neuronal populations on a fast time scale, high-speed calcium imaging will facilitate optical studies of information processing in brain microcircuits. Source: Benjamin F. Grewe and Dominik Langer and Hansjörg Kasper and Björn M. Kampa and Fritjof Helmchen. (2010) High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nature Methods 2(7):399-405.
34 Understanding the biophysical properties and functional organization of single neurons and how they process information is fundamental for understanding how the brain works. The primary function of any nerve cell is to process electrical signals, usually from multiple sources. Electrical properties of neuronal processes are extraordinarily complex, dynamic, and, in the general case, impossible to predict in the absence of detailed measurements. To obtain such a measurement one would, ideally, like to be able to monitor, at multiple sites, subthreshold events as they travel from the sites of origin on neuronal processes and summate at particular locations to influence action potential initiation. This goal has not been achieved in any neuron due to technical limitations of measurements that employ electrodes. To overcome this drawback, it is highly desirable to complement the patch-electrode approach with imaging techniques that permit extensive parallel recordings from all parts of a neuron. Here, we describe such a technique - optical recording of membrane potential transients with organic voltage-sensitive dyes (V(m)-imaging) - characterized by sub-millisecond and sub-micrometer resolution. Our method is based on pioneering work on voltage-sensitive molecular probes. Many aspects of the initial technology have been continuously improved over several decades. Additionally, previous work documented two essential characteristics of V(m)-imaging. Firstly, fluorescence signals are linearly proportional to membrane potential over the entire physiological range (-100 mV to +100 mV). Secondly, loading neurons with the voltage-sensitive dye used here (JPW 3028) does not have detectable pharmacological effects. The recorded broadening of the spike during dye loading is completely reversible. Additionally, experimental evidence shows that it is possible to obtain a significant number (up to hundreds) of recordings prior to any detectable phototoxic effects. At present, we take advantage of the superb brightness and stability of a laser light source at near-optimal wavelength to maximize the sensitivity of the V(m)-imaging technique. The current sensitivity permits multiple site optical recordings of V(m) transients from all parts of a neuron, including axons and axon collaterals, terminal dendritic branches, and individual dendritic spines. The acquired information on signal interactions can be analyzed quantitatively as well as directly visualized in the form of a movie. Popovic M. and Gao X. and Zecevic D. (2012) Voltage-sensitive dye recording from axons, dendrites and dendritic spines of individual neurons in brain slices. Journal of Visualized Experiments 29(69):e4261.
35 There is a pressing need in neuroscience for genetically-encoded, fluorescent voltage probes that can be targeted to specific neurons and circuits to allow study of neural activity using fluorescent imaging. We created 90 constructs in which the voltage sensing portion (S1-S4) of Ciona intestinalis voltage sensitive phosphatase (CiVSP) was fused to circularly permuted eGFP. This led to ElectricPk, a probe that is an order of magnitude faster (taus ~1-2 ms) than any currently published fluorescent protein-based voltage probe. ElectricPk can follow the rise and fall of neuronal action potentials with a modest decrease in fluorescence intensity (~0.7% DF/F). The probe has a nearly linear fluorescence/membrane potential response to both hyperpolarizing and depolarizing steps. This is the first probe based on CiVSP that captures the rapid movements of the voltage sensor, suggesting that voltage probes designed with circularly permuted fluorescent proteins may have some advantages. Barnett, L. and Platisa, J. and Popovic, M. and Pieribone, V.A. and Hughes, T. (2012) A fluorescent, optogenetic voltage sensor capable of resolving action potentials. PloS ONE 7(9):e43454.
36 Silicon bicarbide (SiC) is chemically inert, stable, biocompatible, and has been used to synthesize nanoparticles with cubic symmetry (3C-SiC). In a recent synthesis, 3C-SiC quantum dots with size distribution of ~0.5-3.5 nanometers and emission wavelengths in the UV-visible region were produced. This seminal work has provided a new material for the fabrication of quantum dots, which did not need an added shell for core protection, and were used directly for live cell imaging. (Source: Page 473 in [161])
37 Under many circumstances, the photoluminescence of QDs is drastically quenched through numerous mechanisms. This seemingly undesirable phenomenon can serve as an advantageous feature. For example, quenching mechanism can be designed to act like a molecular switch for fluorescent signals, which would make QDs an ideal homogeneous sensing platform for studying molecular interactions and detecting specific targets. (Source: Page 642 of [174])
38 Multicolor barcoding has enabled high degree of multiplexing in biomolecular assays. Han et al [63] created a two-parameter optical barcode by embedding QDs in polystyrene microbeads in a controlled manner. Both the colors and intensities were precisely modulated for coding. A specific color-intensity combination would pinpoint an exact barcode in the pool of thousands. Theoretically, the combination of m colors and n intensity levels could generate (nm - 1) barcodes. For example, a coding system with 6 colors and 10 intensity levels could have a theoretical coding capacity close to one million. In reality, the coding capacity is typically far less than the theoretical limit due to the spectral cross talk and intensity variations. (Source: Page 644 of [174])
39 Not to be confused with the use of the term in genome sequencing: Depth in DNA sequencing refers to the number of times a nucleotide is read during the sequencing process. Deep sequencing indicates that the coverage, or depth, of the process is many times larger than the length of the sequence under study. (source)
40 Fluorescent semiconductor quantum dots (QDs) can act as energy donors or acceptors with a wide variety of environmentally-sensitive molecules. Conjugation of a single QD to a select number of the selected molecule can optimize the range of sensitivity for a given application, and the relatively large size of the QDs allows them to be tracked individually in cells. Using QDs as FRET acceptors, we have created first-generation sensors for membrane potential which shows good signal to noise and time resolution, but prohibitive toxicity. The challenges of delivery, calibration, and toxicity and plans for improvement of the sensors are presented, in the context of the eventual aim of monitoring membrane potential in a cultured motor neuron model of amyotrophic lateral sclerosis. Source: Quantum dot-FRET systems for imaging of neuronal action potentials. Nadeau, J.L. and Clarke, S.J. and Hollmann, C.A. and Bahcheli, D.M.Proceedings of IEEE Engineering in Medicince & Biology Society Conference. 1:855-8. (2006).
41 Cadmium ions have been shown to bind to thiol groups on critical molecules in the mitochondria and cause enough stress and damage to cause significant cell death.
42 Here’s a tale of hubris in which a back-of-the-envelope estimate was off by enough to make a difference:
A realtor once showed us a house in Morgan Hill, CA and told us that the square footage was 5,347 square feet. I was dubious and I decided to calculate my own estimate working from Google Map imagery. As a basic unit of measure, I used the height of the door frames that were photographed more or less straight on and so I used the height as a scaled 80 inch ruler. Then I aligned this ruler with each of the two axes of the house to obtain overall measurements of the house. I increased these rough estimates by about 10% to account for any foreshortening or inaccuracies in my drawing and that got me 80 × 40 or 3,200 square feet. Finally, I added 500 feet for the upstairs rooms and another 300 square feet for the parts of the first floor on the back that didn’t fit into my rectangle to get 4K. I told the realtor that if I was in the ball park, this would seem to indicate that they included the 1,100 square-foot area of an unfinished outbuilding in their assessment. I told him further that it was possible I could be off by as much as 25% but unlikely.
That night it occurred to me that it would be pretty easy to automate such estimations, and sure enough in the morning morning when I ran a search for “area calculator Google Maps” I found a bunch of interesting apps. The first one I tried was pretty clunky but I thought the second one was well executed. I expect it might turn out to be a pretty handy tool for realtors, surveyors and home buyers. You can access it here. You just type the address of the house or any piece of land and follow the instructions to create a polygonal region that circumscribes the house or land. The corresponding area is then supplied in the output window in a variety of standard units. I’ve attached a screen shot of the region and output for my query. You’ll note that the estimated area is 4,237 square feet which if you assume the second floor is 800 square feet gets you pretty close to the listed 5,479 square feed. So I had to eat crow and admit my initial estimate indeed could have been off by as much as 25%.
  |
43 Apropos the topic of inference involving large amounts of neural-state data, David Heckerman mentioned that there was an IPAM (Institute for Pure & Applied Mathematics) workshop at UCLA in March with goal of facilitating cross fertilization of ideas among leading international thinkers drawn from the disciplines of neuroimaging and computational neuroscience, mathematics, statistics, modeling, and machine learning. Theory, neuroscientific and clinical application perspectives as well as the brain computer interfacing point of view will be discussed.
44 Near-infrared spectroscopy (NIRS) is a spectroscopic method that uses the near-infrared region of the electromagnetic spectrum (from about 800 nm to 2500 nm). Typical applications include pharmaceutical, medical diagnostics (including blood sugar and pulse oximetry), food and agrochemical quality control, and combustion research, as well as research in functional neuroimaging and brain-computer interfaces for medical prosthetics. (source)
45 A scanning electron microscope (SEM) is a type of electron microscope that produces images of a sample by scanning it with a focused beam of electrons. The electrons interact with electrons in the sample, producing various signals that can be detected and that contain information about the sample’s surface topography and composition. The electron beam is generally scanned in a raster scan pattern, and the beam’s position is combined with the detected signal to produce an image. SEM can achieve resolution better than 1 nanometer. (source)
46 Here’s the abstract for Wang et al [163]:
In this study, we develop a novel photoacoustic [microscopy] technique based on gold nanorods (AuNRs) for quantitatively monitoring focused-ultrasound (FUS) induced blood-brain barrier (BBB) opening in a rat model in vivo. This study takes advantage of the strong near-infrared absorption (peak at ~800 nm) of AuNRs and the extravasation tendency from BBB opening foci due to their nano-scale size to passively label the BBB disruption area. Experimental results show that AuNR contrast-enhanced photoacoustic microscopy successfully reveals the spatial distribution and temporal response of BBB disruption area in the rat brains. The quantitative measurement of contrast enhancement has potential to estimate the local concentration of AuNRs and even the dosage of therapeutic molecules when AuNRs are further used as nano-carrier for drug delivery or photothermal therapy. The photoacoustic results also provide complementary information to MRI, being helpful to discover more details about FUS induced BBB opening in small animal models.
47 Gold-nanorods (AuNRs) are “rod-shaped gold nanoparticles with excellent biological compatibility due to their gold surface. They have been applied to photothermal therapy of squamous cell carcinoma in mice25 and have also been used as a nanocarrier for remote control of localized gene expression.” Here’s the abstract for Chen et al [32] which is the paper cited for the work on the application of AuNRs to localized gene expression:
Gold nanorods were attached to the gene of enhanced green fluorescence protein (EGFP) for the remote control of gene expression in living cells. The UV-vis spectroscopy, electrophoresis, and transmission electron microscopy (TEM) were used to study the optical and structural properties of the EGFP DNA and gold nanorod (EGFP-GNR) conjugates before and after femto-second near-infrared (NIR) laser irradiation. Upon NIR irradiation, the gold nanorods of EGFP-GNR conjugates underwent shape transformation that resulted in the release of EGFP DNA. When EGFP-GNR conjugates were delivered to cultured HeLa cells, induced GFP expression was specifically observed in cells that were locally exposed to NIR irradiation. Our results demonstrate the feasibility of using gold nanorods and NIR irradiation as means of remote control of gene expression in specific cells. This approach has potential applications in biological and medical studies.
48 By tuning the averaged dimensions of AuNRs to 40 nm by 10 nm, their absorption peak was shifted to 800-nm wavelength. In addition, polyethylene glycol (PEG) was coated on the surface of the AuNRs to increase their biocompatibility, stealth effect to the immune system, and consequently the circulation time in blood stream.
49 A tunable laser system provided laser pulses with 10-Hz pulse-repetition frequency (PRF), 6.5-ns pulse width, and 800-nm wavelength. In addition to avoiding the strong interference from blood, 800-nm wavelength is also an isosbestic point in absorption spectrum of hemoglobin that we can ignore the effects of blood oxygenation on photoacoustic-microscopy measurements. The laser light was aligned to be confocal with a 25-MHz focused ultrasonic transducer (-6 dB fractional bandwidth: 55%, focal length: 13 mm, v324, Olympus) at 3 mm under the surface of rat brains.
50 Here is the prologue to the description of the Stanford clinical trial for patients with painful bone metastases found on the Pauly lab web site:
Investigators throughout the world are currently applying MR-guided focused ultrasound (MRgFUS) for non-invasive treatments of a variety of diseases and disorders. Typically, focused ultrasound uses a large area ultrasound transducer array outside the body, focused either geometrically or electronically, to a point within the body. The amplification provided by focusing (which can be on the order of 1000-fold) provides the means to generate significant ultrasound intensities deep within the body, with insignificant ultrasound intensities in the intervening tissue. Current clinical systems for body applications are made by InSightec, who makes a variety of systems that are integrated into General Electric MR-scanners, and Phillips, who has a system integrated into their own scanners. An alternative is to use an ultrasound transducer within the target organ or tissue, with little or no focusing, an example of which is the Profound Medical transurethral ultrasound device for prostate treatments, which is currently in clinical trials on Siemens scanners. For brain applications, InSightec has a hemisphere array, which is currently in clinical trials, and Supersonic Imagine is developing a similar system, which will soon begin clinical trials on Siemens scanners.
51 In a tutorial by Gail Haar, she gave some ball-park estimates relating to energy absorption and dissipation: diagnostic scan ~.02 W — scattered energy, and surgical ultrasound ~200 W — energy absorption (source).
52 Several modes of ultrasound are used in medical imaging. In In B-mode (brightness mode) ultrasound, a linear array of transducers simultaneously scans a plane through the body that can be viewed as a two-dimensional image on screen. More commonly known as 2D mode now.
53 From the 2008 Siemens press release announcing the DARPA contract to develop a “Combat Ultrasound Hemorrhage Device”:
The cuff is designed to limit blood loss from penetrating wounds to limbs in fast and slow bleeders, significantly reducing the risk of limb loss and death resulting from irreversible hemorrhagic shock. Once applied to the limb, Siemens Silicon Ultrasound technology within the cuff automatically detects the location and severity of the bleeding within the limb. This triggers therapeutic ultrasound elements within the cuff to emit and focus high-power energy toward the bleeding sites, speeding coagulation and halting bleeding at the injury site. The device is intended for use by minimally-trained operators, curtailing bleeding in a minimal amount of time with automatic treatment and power shut-off.
54 ESB: For ultrasound neural recording: assume you want to get to the whole brain. An analysis:
A key question is whether the ultrasound is used to image, or simply to report bulk activity, which then needs to be sorted out by frequency or temporal code, if you want to resolve individual cells. For the latter, you only get a few channels, as Akram noted. Even with 100 MHz ultrasound, you still get 1 million neurons tops, since you need to use 1kHz of bandwidth each. A more subtle issue is: how do you guarantee that all the probes that bind to a given neuron, will share the same frequency? I can’t think of a good way to do this. Maybe if genetic barcoding could bias the frequency of the acoustic nano-reporter?
If imaging is used, then in principle you could see many more sites than with bulk-activity reporting. Imaging also removes the need for the neuron-address-assignment problem above. An issue that arises is depth of imaging of the brain: even 50 MHz ultrasound will attenuate at 5 dB/cm — see here and here, which is not so bad. 50 MHz means a wavelength in water of 30 microns, so we’re almost, but not quite, there — 1 MHz = 1.5 mm since speed of sound in water is 1500 m/s. Of course, 5 dB /cm isn’t so bad. 0.5 cm mouse brain –> 2.5 dB, which means half the power.
In conclusion, we would want to use 100 MHz ultrasound to image with 15 micron resolution. We might expect to lose 75% of our power, but not that bad. Imaging is better than bulk readout because of the channel sharing problem with bulk readout. We would need a photoacoustic or acoustoacoustic readout transducer.
For imaging:
How might we improve resolution to single cell level? Resolution is not so good. For 1 micron, we need 1.5 GHz. Do acoustic microscopes exist? See here, here and here for prior work.
Can we improve this by thinking more like optics engineers? e.g., how well does "sound-field microscopy" work? What happens if we build incredibly dense microphone arrays, akin to cameras. Could be easy to make if we can get the nanoreporter to work. (But what if sound perturbs neurons? What levels of power would we need?)
Is there such a thing as “super-resolution acoustic microscopy”? Need switchable acoustic reporters. One sound wave toggles, second reads out. Couldn’t find any info about this. Would probably be slow though.
Conclusion of (ii) and (iii) above: what is missing from acoustic microscopy, are there functional contrast agents. Once we have those, it will be easy to build the other hardware. Can we make acoustic calcium sensors? Acoustic voltage sensors? Problem: changing acoustic impedance fast is difficult, much harder than changing optical responses — which are easy to tweak. Cause two particles to move together? Alter stiffness of a bubble? This is the key problem in a way.
Photoacoustic is simple and maybe worth considering too. But what energies will we need? Is light-in, sound-out, fundamentally lower resolution than sound-in, sound-out?
55 Here’s the abstract for the review paper [162] in Science by Wang and Hu on photoacoustic imaging:
Photoacoustic tomography (PAT) can create multiscale multicontrast images of living biological structures ranging from organelles to organs. This emerging technology overcomes the high degree of scattering of optical photons in biological tissue by making use of the photoacoustic effect. Light absorption by molecules creates a thermally induced pressure jump that launches ultrasonic waves, which are received by acoustic detectors to form images. Different implementations of PAT allow the spatial resolution to be scaled with the desired imaging depth in tissue while a high depth-to-resolution ratio is maintained. As a rule of thumb, the achievable spatial resolution is on the order of 1/200 of the desired imaging depth, which can reach up to 7 centimeters. PAT provides anatomical, functional, metabolic, molecular, and genetic contrasts of vasculature, hemodynamics, oxygen metabolism, biomarkers, and gene expression. We review the state of the art of PAT for both biological and clinical studies and discuss future prospects.
56 Radiation in the UV band or higher have enough photon energy to produce ionisation — a process where one or more electrons are removed from a neutral atom by radiation — leading to destruction or modification of living cells. In the case of living tissue, we are primarily interested in the amount of energy deposited in or absorbed by a cell. The gray (Gy), which has units of (joules/kilogram), is the SI unit of absorbed dose, and it is the amount of radiation required to deposit one joule of energy in one kilogram of any kind of matter.
57 The loss tangent is a parameter of a dielectric material that quantifies its inherent dissipation of electromagnetic energy.
58 It might be worth the exercise to consider the complications that arise when we consider modeling the brain as a saline solution and the transmission and reflectance properties of the cranium to determine if we can ignore these factors in making approximations.
59 The near field (or near-field) and far field (or far-field) and the transition zone are regions of time varying electromagnetic field around any object that serves as a source for the field. The different terms for these regions describe the way characteristics of an electromagnetic (EM) field change with distance from the charges and currents in the object that are the sources of the changing EM field. The more distant parts of the far-field are identified with classical electromagnetic radiation.
The basic reason an EM field changes in character with distance from its source is that Maxwell’s equations prescribe different behaviors for each of the two source-terms of electric fields and also the two source-terms for magnetic fields. Electric fields produced by charge distributions have a different character than those produced by changing magnetic fields. Similarly, Maxwell’s equations show a differing behavior for the magnetic fields produced by electric currents, versus magnetic fields produced by changing electric fields. For these reasons, in the region very close to currents and charge-separations, the EM field is dominated by electric and magnetic components produced directly by currents and charge-separations, and these effects together produce the EM “near field.” However, at distances far from charge-separations and currents, the EM field becomes dominated by the electric and magnetic fields indirectly produced by the change in the other type of field, and thus the EM field is no longer affected (or much affected) by the charges and currents at the EM source. (source)
60 Here is a transverse-plane MRI image for a patient with a subdural hematoma plainly showing the pooled blood prior to drainage. The top two plots on the right display the signature pattern associated with such trauma, and demonstrate the diagnostic potential of analyzing eye-tracking data to reliably predict a range of problems from tumors to concussions using technology cheap enough that every family practitioner or contact-sport team physician can afford:
|
|
61 Ed added the following notes:
Ultrasound plus piezoelectric microbubbles: key is to have some way of triggering the change in microbubble physical properties, from a very small and very brief voltage — without this being triggered by noise, of course. Difficult problem. Set up a phase transition on a hair-trigger, perhaps?
In cell culture, can we screen for endogenous ultrasound-to-voltage converting molecules? In human tissue samples?
1 MHz ultrasound is about 1 mm wavelength; to get very accurate measurements, we may need to go to 100 MHZ or higher — at this frequency we start to get significant heating losses in brain tissue, and huge losses in skull. But it seems that we will need to go to high frequencies.
62 The concern was primarily an issue of the power required to penetrate the skull, possible damage to the skull, dura or neural tissues, and reduced resolving power due to scattering. We briefly discussed transcranial MRI-guided focused-ultrasound surgery of brain tumors, an application of ultrasound which, if found safe and effective, will likely spur additional development of ultrasound techniques, perhaps eventually eliminating the reliance on bulky, expensive MRI devices by employing ultrasound or photoacoustic technologies for surgical guidance as well as tumor ablation.
63 As Ed mentioned, green fluorescent proteins (GFPs) dominate in the field. I did discover a few papers on a tagged red-fluorescent channelrhodopsin variant using the ChEF-tdTomato protein including this one. I may have misremembered that adjective “red-white” since the term doesn’t make any sense to me now. I didn’t find any relevant papers with authors from Canadian labs, but did run across this review paper on optogenetics in which Ed’s work and the work of Karl Deisseroth’s lab where Ed worked while at Stanford were featured.
64 This article on phase-change in perfluorocarbon (PFC) droplets provides some insight into the manufacturing process and this article discuses how how this property might be applied in photoacoustic imaging. I also ran across several articles with intriguing titles and abstracts including one on photoacoustic imaging with contrast agents and another on photoacoustic tomography
65 The Janelia Farm Research Campus of HHMI features small, cross-disciplinary teams that bring together “chemists, physicists, computational scientists, and engineers into close collaboration with biologists. Janelia collaborations have yielded several new technologies in optical imaging and genetic sequencing, and in using software to assemble sophisticated 3D models of neural circuitry.” Loren Looger’s team at Janelia developed the calcium imaging technology that Ed mentioned:
The most in demand of any of the tools he has developed, that indicator, GCaMP3, has been distributed to hundreds of labs where it illuminates neural activity that went unnoticed with earlier sensors. Still, neuroscientists are demanding a suite of similar tools that excel at different aspects of calcium sensing, so the overall effort to build better genetically encoded calcium indicators has, like GCaMP3, spread beyond Looger’s lab. Thanks to a large-scale push to generate and evaluate new versions of the protein, GCaMP3 has been mostly superseded by GCaMP5, which produces even less background fluorescence, gives a greater signal in the presence of calcium, and picks up more activity in the brains of living animals. (source)
66 From the Materials Project description out of Gerbrand Cedars’s Lab at MIT:
-
It takes an average of 20 years to bring new materials from lab to commercialization, and new materials are often discovered through a mix of intuition and serendipity. This development process makes it difficult to bring new materials needed urgently (like better Li-ion batteries, solar photovoltaics, or thermoelectrics) to market in a reasonable time.
The goal of the Materials Project is to accelerate the materials discovery and design process by (a) computing the fundamental properties of all known inorganic compounds and (b) helping researchers predict new materials with desired properties. Such information would allow researchers to rationally design new materials, dramatically cutting discovery and optimization times. The Materials Project includes aspects of computer science (software design, web design, database design), high-performance computing, theoretical physics (accurately modeling diverse chemical spaces), and materials science (linking fundamental physics to device properties).
67 A molecule or compound is made when two or more atoms form a chemical bond, linking them together. The two types of bonds are ionic bonds and covalent bonds. In an ionic bond, the atoms are bound together by the attraction between oppositely-charged ions. For example, sodium and chloride form an ionic bond, to make NaCl, or table salt. In a covalent bond, the atoms are bound by shared electrons. If the electron is shared equally between the atoms forming a covalent bond, then the bond is said to be nonpolar. Usually, an electron is more attracted to one atom than to another, forming a polar covalent bond. For example, the atoms in water, H2O, are held together by polar covalent bonds (source).
Here are some properties of covalent compounds, which are generally referred to as molecular compounds presumably to emphasize their central role in so many important chemical processes and distinguish them from ionic compounds (source):
Most covalent compounds have relatively low melting points and boiling points. While the ions in an ionic compound are strongly attracted to each other, covalent bonds create molecules that can separate from each other when a lower amount of energy is added to them. Therefore, molecular compounds usually have low melting and boiling points.
Covalent compounds usually have lower enthalpies of fusion and vaporization than ionic compounds. The enthalpy of fusion is the amount of energy needed, at constant pressure, to melt one mole of a solid substance. The enthalpy of vaporization is the amount of energy, at constant pressure, required to vaporize one mole of a liquid. On average, it takes only 1% to 10% as much heat to change the phase of a molecular compound as it does for an ionic compound.
Covalent compounds tend to be soft and relatively flexible. This is largely because covalent bonds are relatively flexible and easy to break. The covalent bonds in molecular compounds cause these compounds to take form as gases, liquids and soft solids. As with many properties, there are exceptions, primarily when molecular compounds assume crystalline forms.
Covalent compounds tend to be more flammable than ionic compounds. Many flammable substances contain hydrogen and carbon atoms which can undergo combustion, a reaction that releases energy when the compound reacts with oxygen to produce carbon dioxide and water. Carbon and hydrogen have comparable electronegativies so they are found together in many molecular compounds.
When dissolved in water, covalent compounds don’t conduct electricity. Ions are needed to conduct electricity in an aqueous solution. Molecular compounds dissolve into molecules rather than dissociate into ions, so they typically do not conduct electricity very well when dissolved in water.
Many covalent compounds don’t dissolve well in water. There are many exceptions to this rule, just as there are many salts (ionic compounds) that don’t dissolve well in water. However, many covalent compounds are polar molecules that do dissolve well in a polar solvent, such as water. Examples of molecular compounds that dissolve well in water are sugar and ethanol. Examples of molecular compounds that don’t dissolve well in water are oil and polymerized plastic.
Here are the properties shared by the ionic compounds. Notice that the properties of ionic compounds relate to how strongly the positive and negative ions attract each other in an ionic bond (source):
Ionic compounds form crystals. Ionic compounds form crystal lattices rather than amorphous solids. Although molecular compounds form crystals, they frequently take other forms plus molecular crystals typically are softer than ionic crystals.
Ionic compounds have high melting points and high boiling points. High temperatures are required to overcome the attraction between the positive and negative ions in ionic compounds. Therefore, a lot of energy is required to melt ionic compounds or cause them to boil.
Ionic compounds have higher enthalpies of fusion and vaporization than molecular compounds. Just as ionic compounds have high melting and boiling points, they usually have enthalpies of fusion and vaporization that may be 10 to 100 times higher than those of most molecular compounds. The enthalpy of fusion is the heat required melt a single mole of a solid under constant pressure. The enthalpy of vaporization is the heat required for vaporize one mole of a liquid compound under constant pressure.
Ionic compounds are hard and brittle. Ionic crystals are hard because the positive and negative ions are strongly attracted to each other and difficult to separate, however, when pressure is applied to an ionic crystal then ions of like charge may be forced closer to each other. The electrostatic repulsion can be enough to split the crystal, which is why ionic solids also are brittle.
Ionic compounds conduct electricity when they are dissolved in water. When ionic compounds are dissolved in water the dissociated ions are free to conduct electric charge through the solution. Molten ionic compounds (molten salts) also conduct electricity.
Ionic solids are good insulators. Although they conduct in molten form or in aqueous solution, ionic solids do not conduct electricity very well because the ions are bound so tightly to each other.
Here are some examples of covalent bonds and covalent compounds. Covalent compounds also are known as molecular compounds. Organic compounds, such as carbohydrates, lipids, proteins and nucleic acids, are all examples of molecular compounds. You can recognize these compounds because they consist of nonmetals bonded to each other: PCl3, CH3CH2OH, O3 — ozone, H2 — hydrogen, H2O — water, HCl — hydrogen chloride, CH4 — methane, NH3 — ammonia, and CO2 — carbon dioxide. (source)
Here are some examples of ionic bonds and ionic compounds. You can recognize ionic compounds because they consist of a metal bonded to a nonmetal: NaBr — sodium bromide, KBr — potassium bromide, NaCl — sodium chloride, NaF — sodium fluoride, KI — potassium iodide, KCl — potassium chloride, CaCl2 — calcium chloride, K2O — potassium oxide, and MgO — magnesium oxide. (source)
You can see the pattern of how covalent and ionic bonds arise in nature in the organization of the periodic table of elements:
 |
68 The extracellular matrix of the adult brain tissue has a unique composition. The striking feature of this matrix is the prominence of lecticans, proteoglycans that contain a lectin domain and a hyaluronic acid-binding domain. Hyaluronic acid and tenascin family adhesive/anti-adhesive proteins are also abundant. Matrix proteins common in other tissues are nearly absent in adult brain. The brain extracellular matrix appears to have trophic effects on neuronal cells and affect neurite outgrowth. The unique composition of this matrix may be responsible for the resistance of brain tissue toward invasion by tumors of non-neuronal origin. — excerpted this review article [132] by Erkki Ruoslahti.
69 I probably misheard Chris or he used different estimates for SEM-voxel size or mouse-brain volume since when I did the calculation I got 400,000TB at one byte per voxel. Here are the steps in my calculation: One voxel from a serial-block-face scanning electron microscope (SBFSEM) is 10 × 10 × 25 nm or 102 * 25 = 2,500 nm3. The volume of a mouse brain is about 103 mm3. A millimeter is a million nanometers, a cubic millimeter is a million-cubed cubic nanometers and so a mouse brain is (10 * 106)3 = 1021 nm3 and a complete scan would span (1021 / 2,500) SBFSEM voxels. A terabyte is 1,099,511,627,776 bytes and so the number of terabyte disks we would need to store a complete scan is (1021 / 2,500) / 1,099,511,627,776) or approximately (1021 / 2,500) / 1012 = 400,000 TB disks.
70 Immunohistochemistry refers to the process of detecting antigens (e.g., proteins) in cells of a tissue section by exploiting the principle of antibodies binding specifically to antigens in biological tissues (source).
71 Confocal point sensor principle from Minsky’s patent (source):
 |
72 To simulate exposure to a carcinogen, the target cells were incubated with a metabolite of a chemical called ben-zo[a]pyrene (BaP), a known cancer-causing environmental agent often found in polluted urban atmospheres.
73 Here is a sample of recent papers describing hybrid imaging techniques that combine ultrasound with various light-based technologies:
Slow light for deep tissue imaging with ultrasound modulation.
Huiliang Zhang, Mahmood Sabooni, Lars Rippe, Chulhong Kim, Stefan Kröll, Lihong V. Wang, and Philip R. Hemmer.
Applied Physics Letters, 100:13, 2012.
Abstract: Slow light has been extensively studied for applications ranging from optical delay lines to single photon quantum storage. Here, we show that the time delay of slow-light significantly improves the performance of the narrow-band spectral filters needed to optically detect ultrasound from deep inside highly scattering tissue. We demonstrate this capability with a 9 cm thick tissue phantom, having 10 cm-1 reduced scattering coefficient, and achieve an unprecedented background-free signal. Based on the data, we project real time imaging at video rates in even thicker phantoms and possibly deep enough into real tissue for clinical applications like early cancer detection.
Breaking the spatial resolution barrier via iterative sound-light interaction in deep tissue microscopy.
Ke Si, Reto Fiolka, Meng Cui.
Science Reports, 2:748, 2012.
Abstract: Optical microscopy has so far been restricted to superficial layers, leaving many important biological questions unanswered. Random scattering causes the ballistic focus, which is conventionally used for image formation, to decay exponentially with depth. Optical imaging beyond the ballistic regime has been demonstrated by hybrid techniques that combine light with the deeper penetration capability of sound waves. Deep inside highly scattering media, the sound focus dimensions restrict the imaging resolutions. Here we show that by iteratively focusing light into an ultrasound focus via phase conjugation, we can fundamentally overcome this resolution barrier in deep tissues and at the same time increase the focus to background ratio. We demonstrate fluorescence microscopy beyond the ballistic regime of light with a threefold improved resolution and a fivefold increase in contrast. This development opens up practical high resolution fluorescence imaging in deep tissues.
Combined ultrasonic and photoacoustic system for deep tissue imaging.
Chulhong Kim, Todd N. Erpelding, Ladislav Jankovic, Lihong V. Wang.
SPIE 7899, Photons Plus Ultrasound: Imaging and Sensing, 2011.
Abstract: A combined ultrasonic and photoacoustic imaging system is presented that is capable of deep tissue imaging. The system consists of a modified clinical ultrasound array system and tunable dye laser pumped by a Nd:YAG laser. The system is designed for noninvasive detection of sentinel lymph nodes and guidance of needle biopsies for axillary lymph node staging in breast cancer patients. Using a fraction of the American National Standards Institute (ANSI) safety limit, photoacoustic imaging of methylene blue achieved penetration depths of greater than 5 cm in chicken breast tissue. Photoacoustic imaging sensitivity was measured by varying the concentration of methylene blue dye placed at a depth of 3 cm within surrounding chicken breast tissue. Signal-to-noise ratio, noise equivalent sensitivity, and axial spatial resolution were quantified versus depth based on in vivo and chicken breast tissue experiments. The system has been demonstrated in vivo for detecting sentinel lymph nodes in rats following intradermal injection of methylene blue. These results highlight the clinical potential of photoacoustic image-guided identification and needle biopsy of sentinel lymph nodes for axillary staging in breast cancer patients.
74 This excerpt taken from a technical report produced by the Society for Nuclear Medicine and Molecular Medicine describes examples of both untargeted and targeted microbubble contrast agents:
The Ultrasound Contrast Mode offers the ability to enhance the contrast of the ultrasound image (untargeted contrast agents) for detecting vascular and myocardial perfusion, and to detect, quantify, and perform dynamic imaging at the molecular level, such as inflammation and angiogenesis (targeted contrast agents).The untargeted contrast agents (micro-bubbles, BRACCO Research SA, Geneva, Switzerland) are composed of a gas mixture (Nitrogen and Perfluorobutane) and are stabilized by a phospholipids monolayer with a median diameter of 2.3 to 2.9 μm.
Targeted microbubbles contain Streptavidin incorporated into the lipid shell which can be conjugated to any biotinylated ligand of choice, i.e. antibodies, proteins, DNA.
For the study of angiogenesis, the microbubbles are functionalized with biotinylated anti-VEGF-R2 monoclonal antibody (FLK1-VEGFR2, eBiosciences, San Diego, CA). A rat isotype control (IgG2A, eBiosciences, San Diego, CA) is used as the control.
Studying inflammation processes, the microbubbles are functionalized with biotinylated anti-mouse CD62P monoclonal antibody (P-Selectin, BD Pharmingen, BD Biosciences, Franklin Lakes, NJ). A rat isotype control (IgG1 Lambda Isotype, eBiosciences, San Diego, CA) is used as the control.
75 Here’s an excerpt from Akram S. Sadek’s description of his NEMS multiplexer inspired by current theories of the cochlea:
[T]he cochlea acts as an inverse Fourier transform — it takes a single input channel that is multiplexed at different frequencies and decomposes it to multiple, individual output channels. In our scheme, we implement the inverse of this. We use a bank of piezoelectric nanoelectromechanical systems (NEMS), beam structures of different length, each resonating via piezoelectric actuation at well separated frequencies. Each NEMS device performs the job of the inner and outer hair cells concurrently. It uses the piezoelectric effect to actuate the devices to resonance, and also harnesses the piezoelectric effect to detect electrical signals applied to the device in a very sensitive fashion.Each NEMS device is attached to the electrical output from a different nanoscale sensor. The signals from the sensors vary the resonance of each device very slightly. It does this by varying the baseline stress in each beam, much like tuning a guitar string by varying the tension. What this means is that if we force each NEMS device to resonate at a fixed frequency, ’tuning’ the natural frequency of each device using the output signals from a nanosensor will cause fluctuations in the amplitude, and thence power of the mechanical resonance of each NEMS device. If this correlation between power fluctuation and sensor signal is linear — which is the case for our system — then the output from a nanosensor can be encoded using those power fluctuations.
Now that each sensor channel has been encoded from the time domain into the frequency domain, we can multiplex the signals in frequency space onto a single output channel. In our scheme, we do this optically by shining a laser on the different resonators concurrently. The mechanical resonance causes power fluctuations in the reflected laser light at different frequencies via interference. This single optical output encodes all the information from the different sensors, and can be decoded to recover the original signal using a reverse process, in our case done using a standard signal processing technique. As each frequency is tied to a specific, known sensor, the system is addressable.
76 Here is an excerpt from Jones [76] in which the author outlines the dynamical characteristics of biological linear motors:
Underlying the mechanism of this remarkable process are those central features of nanoworld, namely Brownian motion, conformational change and stickiness. Both ATP and ADP bind to the same place in the head-group of myosin, but, because these two molecules have slightly different shape, the shape of the myosin molecule itself is distorted in different ways when each molecule is associated with its binding pocket. A consequence of this change in shape is that the degree to which another part of the myosin molecule sticks to the actin filament changes. The hydrolysis of ATP to ADP thus leads to the changes in the shape and stickiness of the myosin which underlie the action of the motor. (source: page 147 in [76])
77 Here is an excerpt from Ventola [157] discussing how nanoparticles — denoted “NP” in the following — are removed from circulation by the immune system; the abbreviation “RES” denotes the reticuloendothelial system which is the part of the immune system consisting of phagocytes located in reticular connective tissue and is referred to as the mononuclear phagocyte system in modern texts:
NPs are generally cleared from circulation by immune system proteins called opsonins, which activate the immune complement system and mark the NPs for destruction by macrophages and other phagocytes. Neutral NPs are opsonized to a lesser extent than charged particles, and hydrophobic particles are cleared from circulation faster than hydrophilic particles. NPs can therefore be designed to be neutral or conjugated with hydrophilic polymers (such as PEG) to prolong circulation time. The bioavailability of liposomal NPs can also be increased by functionalizing them with a PEG coating in order to avoid uptake by the RES. Liposomes functionalized in this way are called “stealth liposomes.”NPs are often covered with a PEG coating as a general means of preventing opsonization, reducing RES uptake, enhancing biocompatibility, and/or increasing circulation time. SPIO NPs can also be made water-soluble if they are coated with a hydrophilic polymer (such as PEG or dextran), or they can be made amphophilic or hydrophobic if they are coated with aliphatic surfactants or liposomes to produce magnetoliposomes. Lipid coatings can also improve the biocompatibility of other particles.
Relevant to elimination by the kidneys, this paper by Choi et al [33] claims to have “precisely defined the requirements for renal filtration and urinary excretion of inorganic, metal-containing nanoparticles”, and, while somewhat narrowly focused, provides some useful general information regarding renal filtration.
78 A complete action-potential cycle takes around 4 milliseconds consisting of about 2 ms for polarization and depolarization of the axon cell membrane followed by a refractory period of about 2 ms during which the neuron is unable to fire.
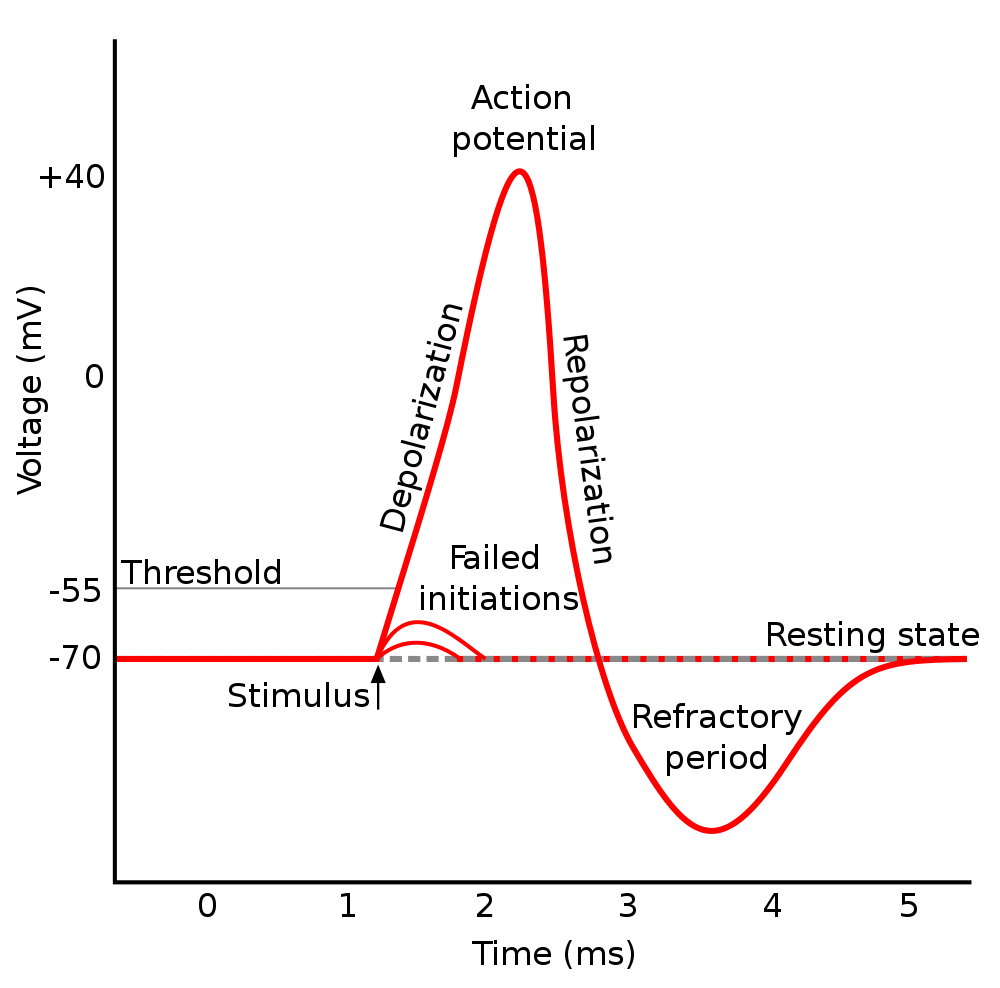 |
79 The Central Dogma of Molecular Biology, first introduced by Francis Crick in 1958, describes flow of genetic information within a biological system: covering replication (DNA to DNA), transcription (DNA to RNA) and translation (RNA to protein) as well as some less well known transfers including DNA methylation which “stably alters the gene expression pattern in cells such that cells can ‘remember where they have been’ or decrease gene expression; for example, cells programmed to be pancreatic islets during embryonic development remain pancreatic islets throughout the life of the organism without continuing signals telling them that they need to remain islets”.
80 In accord with kinetic theory, small particles in a gas or solution are a constant state of motion at all temperatures above absolute zero. The thermal motion of particles rises with the temperature of those particles and is governed by the laws of thermodynamics.
81 Action potentials require the movement of calcium, potassium and sodium ions across the axonal membrane. Once initiated, the action potential propagates down the axon from the soma toward the synaptic termini at at anywhere from 0.5 to 100 meters per second depending on the diameter of the axon and whether or not the axon is myelinated. You can find a good discussion of neural signal propagation here and an instructive animation here.
82 See this Khan Academy video describing a simple organic-chemistry reaction and discussing why some reaction products are more likely than others as predicted by Markovnikov’s rule.
83 This use of the term “barcoding” is different from the method of using the existing DNA of organisms as ready-made barcodes for taxonomic identification. Apparently “barcoding” was first described in technical detail in George M. Church’s Harvard PhD.
84 DNA origami is the “nanoscale folding of DNA to create arbitrary two and three dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material, through design of its base sequences.” (source). A long strand of DNA is programmed to self-assemble by folding in on itself with the help of shorter strands called staples to create predetermined forms.
85 Technically, HGP did not sequence the entire DNA found in human cells. See here for a more careful analysis of what HGP actually accomplished.
86 ENCODE, the Encyclopedia of DNA Elements, is a project funded by the National Human Genome Research Institute to “identify all regions of transcription, transcription factor association, chromatin structure and histone modification in the human genome sequence. Thanks to the identification of these functional elements, 80% of the components of the human genome now have at least one biochemical function associated with them.”
87 Viruses obviously can pass through the blood brain barrier aided at least in some cases by the glial cells that provide nourishment to neurons (source):
Now viruses rank among the environmental factors thought to trigger brain-ravaging diseases such as multiple sclerosis (MS) and Alzheimer’s disease. Human herpesvirus-6 (HHV-6), in particular, has been linked to MS in past studies. Neuroscientist Steven Jacobson and his colleagues at the National Institute of Neurological Disorders and Stroke have determined that the virus makes its entry to the human brain through the olfactory pathway, right along with the odors wafting into our nose.The researchers tested samples of brain cells from people with MS and healthy control subjects and found evidence of the virus in the olfactory bulb in both groups. Infection via the nasal passage is probably quite common, as is harboring a dormant reservoir of HHV-6, but in people with MS, the virus is active. Genetics and other unknown environmental factors probably determine the likelihood of the virus reactivating once inside the brain, which can cause the disease to progress.
The virus appears to invade the brain by infecting a type of glial cell called olfactory ensheathing cells (OECs), which nourish smell-sensing neurons and guide them from the olfactory bulb to their targets in the nervous system. These targets include the limbic system, a group of evolutionarily old structures deep in the brain, “which is where viruses like to reactivate,” Jacobson explains. He points out that olfactory neurons and their OECs are among the few brain cells known to regenerate throughout our life. This neurogenesis may keep our sense of smell sharp, but at the cost of providing the virus the opportunity to spread.
89 Transport rates referred to as axoplasmic transport rates depend on what’s being transported. The axoplasmic transport rate for dimers like actin and tubulin from which microtubules are constructed is about 0.2–4 mm per day. Mitochondrial proteins move somewhat faster at around 15–50 mm per day, while peptides — short polymers of amino acids held together by peptide bonds — and glycolipids — metabolically important lipids with attached carbohydrates — are transported at the blazingly fast speed of 200–400 mm per day.
88 Viruses can be transmitted in one of two directions. First, one must understand the underlying mechanism of axoplasmic transport. Within the axon are long slender protein complexes called microtubules. They act as a cytoskeleton to help the cell maintain its shape. These can also act as highways within the axon and facilitate transport of neurotransmitter-filled vesicles and enzymes back and forth between the cell body, or soma and the axon terminal, or synapse.89
Transport can proceed in either direction: anterograde (from soma to synapse), or retrograde (from synapse to soma). Neurons naturally transport proteins, neurotransmitters, and other macromolecules via these cellular pathways. Neuronal tracers, including viruses, take advantage of these transport mechanisms to distribute a tracer throughout a cell. Researchers can use this to study synaptic circuitry.
Anterograde tracing is the use of a tracer that moves from soma to synapse. Anterograde transport uses a protein called kinesin to move viruses along the axon in the anterograde direction. Retrograde tracing is the use of a tracer that moves from synapse to soma. Retrograde transport uses a protein called dynein to move viruses along the axon in the retrograde direction. It is important to note that different tracers show characteristic affinities for dynein and kinesin, and so will spread at different rates (source).
91 A ribosome is a large and complex molecular machine, found within all living cells, that serves as the primary site of biological protein synthesis. Ribosomes link amino acids together in the order specified by messenger RNA (mRNA) molecules. Ribosomes consist of two major subunits–the small ribosomal subunit reads the mRNA, while the large subunit joins amino acids to form a polypeptide chain. Each subunit is composed of one or more ribosomal RNA (rRNA) molecules and a variety of proteins (source).
90 Scaling laws complicate this process of miniaturization given that “[a]s the sizes got smaller, one would have to redesign some tools, because the relative strength of various forces would change. Although gravity would become unimportant, surface tension would become more important, Van der Waals attraction would become important, etc. Feynman mentioned these scaling issues during his talk. Nobody has yet attempted to implement this thought experiment, although it has been noted that some types of biological enzymes and enzyme complexes (especially ribosomes91) function chemically in a way close to Feynman’s vision.” — excerpted from Wikipedia.
92 Richard Feynman gave his highly influential talk entitled “There’s Plenty of Room at the Bottom” on December 29th 1959 at the annual meeting of the American Physical Society at the California Institute of Technology. It was his vision on how physics and engineering could move in the direction that could eventually create nanotechnology. This talk is a retelling of that early lecture at a point in time when several of the component technologies that he envisioned. Specifically, in the early 1980’s the scanning tunneling microscope was invented at IBM-Zurich in Switzerland. This was the first instrument that was able to “see” atoms. A few years later, the Atomic Force Microscope was invented, expanding the capabilities and types of materials that could be investigated. Hence, Scanning Probe Microscopy was born, and since then multiple similar techniques have evolved from these instruments to “see” different properties at the nanometer scale. In addition, “older” techniques such as electron microscopy have continued to evolve as well, and now can image in the nanometer range. Currently, there are a large number of complementary instruments that help scientists in the nano realm.
93 Identifying the connection network directly from the tissue structure is practically impossible, even in cell cultures with only a few thousand neurons. In contrast, there are well-developed methods for recording dynamic neuronal activity patterns. Such patterns indicate which neuron transmitted a signal at what time, making them a kind of neuronal conversation log.
The Gottingen-based team headed by Theo Geisel, Director at the Max Planck Institute for Dynamics and Self-Organization, has now made use of these activity patterns. The scientists use data from calcium fluorescence measurements that were recorded in collaboration with the University of Barcelona.
This imaging method uses specially tailored molecules placed in a cell that fluoresce when they bind calcium. Since the calcium concentration inside a neuron follows its electrical activity, it is possible to record the activity of thousands of neurons simultaneously in a cell culture or in the living brain.
However,the speed of the communication is too high to directly observe directly how an impulse is “fired” and thus tease apart whether a connection is direct or takes places across several stations. By taking into account these difficulties, the algorithm developed in Geisel’s team makes it possible to obtain from the measured data remarkably precise information about the lines of connection in the neural network.
94 The article states that the “key novelty in the atlas is the mapping of microscopic features (such as average cell size and packing density) within the white matter, which contains the neuronal fibers that transmit information around the living brain. The results of the project provide new depth and accuracy in our understanding of the human brain in health and disease.” and that the atlas “describes the brain’s microstructure in standardized space, which enables non-expert users, such as physicians or medical researchers, to exploit the wealth of knowledge it contains. The atlas contains a variety of new images that represent different microscopic tissue characteristics, such as the fiber diameter and fiber density across the brain, all estimated using MRI. These images will serve as the reference standard of future brain studies in both medicine and basic neuroscience.”
95 Viruses obviously can pass through the blood brain barrier aided at least in some cases by the glial cells that provide nourishment to neurons (source):
Now viruses rank among the environmental factors thought to trigger brain-ravaging diseases such as multiple sclerosis (MS) and Alzheimer’s disease. Human herpesvirus-6 (HHV-6), in particular, has been linked to MS in past studies. Neuroscientist Steven Jacobson and his colleagues at the National Institute of Neurological Disorders and Stroke have determined that the virus makes its entry to the human brain through the olfactory pathway, right along with the odors wafting into our nose.The researchers tested samples of brain cells from people with MS and healthy control subjects and found evidence of the virus in the olfactory bulb in both groups. Infection via the nasal passage is probably quite common, as is harboring a dormant reservoir of HHV-6, but in people with MS, the virus is active. Genetics and other unknown environmental factors probably determine the likelihood of the virus reactivating once inside the brain, which can cause the disease to progress.
The virus appears to invade the brain by infecting a type of glial cell called olfactory ensheathing cells (OECs), which nourish smell-sensing neurons and guide them from the olfactory bulb to their targets in the nervous system. These targets include the limbic system, a group of evolutionarily old structures deep in the brain, “which is where viruses like to reactivate,” Jacobson explains. He points out that olfactory neurons and their OECs are among the few brain cells known to regenerate throughout our life. This neurogenesis may keep our sense of smell sharp, but at the cost of providing the virus the opportunity to spread.
96 Viruses can be transmitted in one of two directions. First, one must understand the underlying mechanism of axoplasmic transport. Within the axon are long slender protein complexes called microtubules. They act as a cytoskeleton to help the cell maintain its shape. These can also act as highways within the axon and facilitate transport of neurotransmitter-filled vesicles and enzymes back and forth between the cell body, or soma and the axon terminal, or synapse. Transport can proceed in either direction: anterograde (from soma to synapse), or retrograde (from synapse to soma). Neurons naturally transport proteins, neurotransmitters, and other macromolecules via these cellular pathways. Neuronal tracers, including viruses, take advantage of these transport mechanisms to distribute a tracer throughout a cell. Researchers can use this to study synaptic circuitry.
Anterograde tracing is the use of a tracer that moves from soma to synapse. Anterograde transport uses a protein called kinesin to move viruses along the axon in the anterograde direction. Retrograde tracing is the use of a tracer that moves from synapse to soma. Retrograde transport uses a protein called dynein to move viruses along the axon in the retrograde direction. It is important to note that different tracers show characteristic affinities for dynein and kinesin, and so will spread at different rates (source).
98 A ribosome is a large and complex molecular machine, found within all living cells, that serves as the primary site of biological protein synthesis. Ribosomes link amino acids together in the order specified by messenger RNA (mRNA) molecules. Ribosomes consist of two major subunits–the small ribosomal subunit reads the mRNA, while the large subunit joins amino acids to form a polypeptide chain. Each subunit is composed of one or more ribosomal RNA (rRNA) molecules and a variety of proteins (source).
97 Scaling laws complicate this process of miniaturization given that “[a]s the sizes got smaller, one would have to redesign some tools, because the relative strength of various forces would change. Although gravity would become unimportant, surface tension would become more important, Van der Waals attraction would become important, etc. Feynman mentioned these scaling issues during his talk. Nobody has yet attempted to implement this thought experiment, although it has been noted that some types of biological enzymes and enzyme complexes (especially ribosomes98) function chemically in a way close to Feynman’s vision.” — excerpted from Wikipedia.
99 White matter corresponds to myelinated axons of certain cortical neurons. The distribution of white and gray matter shifts rather dramatically as we age. Early in development most processing is local in nature but as we mature we engage more functional areas of the brain and thus processing is facilitated by accelerating communication between these distant areas. The sheath that surrounds myelinated axons corresponds to the bodies of Schwann cells and provides a degree of insulation that speeds transmission and reduces crosstalk. Between the Schwann cells are the nodes of Ranvier which are exposed to the cytoplasm. Most of the ion channels on myelinated axons are located at the nodes. The following graphic shows how the normalized ratio of gray and White matter changes as we age (source):
 |
100 Christof Koch estimates the total number of neurons in the cerebral cortex at 20 billion and the total number of synapses in the cerebral cortex at 240 trillion — Biophysics of Computation. Information Processing in Single Neurons, New York: Oxford Univ. Press, 1999, Page 87.
101 It is estimated that we lose 80,000-100,000 neurons each day as part of normal, programmed cell death, According to our current understanding most of these are replaced by new, migrating neurons made by the hippocampus. Of course, with Alzheimer’s Disease, Parkinson’s Disease, and Dementia, the loss of neurons becomes significant, estimated in the range 400,000-800,000 per day. During the fetal period up to birth, about one-third to two-third of all neurons made during the embryonic period die as cells migrate and differentiate. By the age of twenty, a person will have lost about one third of what is left after the massive neuronal deaths occurring during birth.
102 The conduction velocity of myelinated neurons varies roughly linearly with axon diameter whereas the speed of unmyelinated neurons varies roughly as the square root (source).
103 Richard Feynman gave his highly influential talk entitled “There’s Plenty of Room at the Bottom” on December 29th 1959 at the annual meeting of the American Physical Society at the California Institute of Technology. It was his vision on how physics and engineering could move in the direction that could eventually create nanotechnology. This talk is a retelling of that early lecture at a point in time when several of the component technologies that he envisioned. Specifically, in the early 1980’s the scanning tunneling microscope was invented at IBM-Zurich in Switzerland. This was the first instrument that was able to “see” atoms. A few years later, the Atomic Force Microscope was invented, expanding the capabilities and types of materials that could be investigated. Hence, Scanning Probe Microscopy was born, and since then multiple similar techniques have evolved from these instruments to “see” different properties at the nanometer scale. In addition, “older” techniques such as electron microscopy have continued to evolve as well, and now can image in the nanometer range. Currently, there are a large number of complementary instruments that help scientists in the nano realm.
104 The article states that the “key novelty in the atlas is the mapping of microscopic features (such as average cell size and packing density) within the white matter, which contains the neuronal fibers that transmit information around the living brain. The results of the project provide new depth and accuracy in our understanding of the human brain in health and disease.” and that the atlas “describes the brain’s microstructure in standardized space, which enables non-expert users, such as physicians or medical researchers, to exploit the wealth of knowledge it contains. The atlas contains a variety of new images that represent different microscopic tissue characteristics, such as the fiber diameter and fiber density across the brain, all estimated using MRI. These images will serve as the reference standard of future brain studies in both medicine and basic neuroscience.”
105 Identifying the connection network directly from the tissue structure is practically impossible, even in cell cultures with only a few thousand neurons. In contrast, there are well-developed methods for recording dynamic neuronal activity patterns. Such patterns indicate which neuron transmitted a signal at what time, making them a kind of neuronal conversation log.
The Gottingen-based team headed by Theo Geisel, Director at the Max Planck Institute for Dynamics and Self-Organization, has now made use of these activity patterns. The scientists use data from calcium fluorescence measurements that were recorded in collaboration with the University of Barcelona.
This imaging method uses specially tailored molecules placed in a cell that fluoresce when they bind calcium. Since the calcium concentration inside a neuron follows its electrical activity, it is possible to record the activity of thousands of neurons simultaneously in a cell culture or in the living brain.
However,the speed of the communication is too high to directly observe directly how an impulse is “fired” and thus tease apart whether a connection is direct or takes places across several stations. By taking into account these difficulties, the algorithm developed in Geisel’s team makes it possible to obtain from the measured data remarkably precise information about the lines of connection in the neural network.