seaborn.residplot#
- seaborn.residplot(data=None, *, x=None, y=None, x_partial=None, y_partial=None, lowess=False, order=1, robust=False, dropna=True, label=None, color=None, scatter_kws=None, line_kws=None, ax=None)#
Plot the residuals of a linear regression.
This function will regress y on x (possibly as a robust or polynomial regression) and then draw a scatterplot of the residuals. You can optionally fit a lowess smoother to the residual plot, which can help in determining if there is structure to the residuals.
- Parameters:
- dataDataFrame, optional
DataFrame to use if
xandyare column names.- xvector or string
Data or column name in
datafor the predictor variable.- yvector or string
Data or column name in
datafor the response variable.- {x, y}_partialvectors or string(s) , optional
These variables are treated as confounding and are removed from the
xoryvariables before plotting.- lowessboolean, optional
Fit a lowess smoother to the residual scatterplot.
- orderint, optional
Order of the polynomial to fit when calculating the residuals.
- robustboolean, optional
Fit a robust linear regression when calculating the residuals.
- dropnaboolean, optional
If True, ignore observations with missing data when fitting and plotting.
- labelstring, optional
Label that will be used in any plot legends.
- colormatplotlib color, optional
Color to use for all elements of the plot.
- {scatter, line}_kwsdictionaries, optional
Additional keyword arguments passed to scatter() and plot() for drawing the components of the plot.
- axmatplotlib axis, optional
Plot into this axis, otherwise grab the current axis or make a new one if not existing.
- Returns:
- ax: matplotlib axes
Axes with the regression plot.
See also
regplotPlot a simple linear regression model.
jointplotDraw a
residplot()with univariate marginal distributions (when used withkind="resid").
Examples
Pass
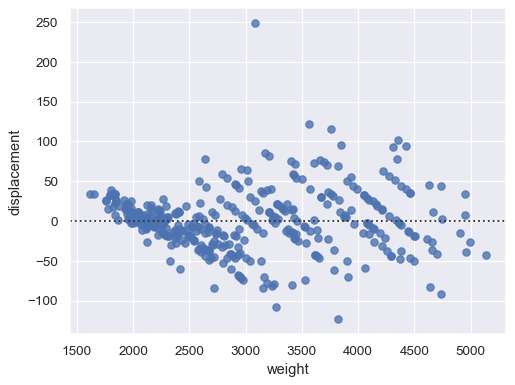
xandyto see a scatter plot of the residuals after fitting a simple regression model:sns.residplot(data=mpg, x="weight", y="displacement")
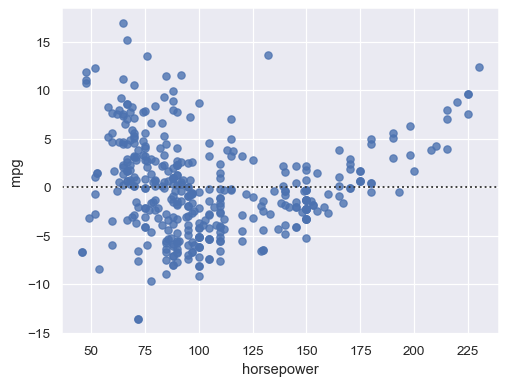
Structure in the residual plot can reveal a violation of linear regression assumptions:
sns.residplot(data=mpg, x="horsepower", y="mpg")
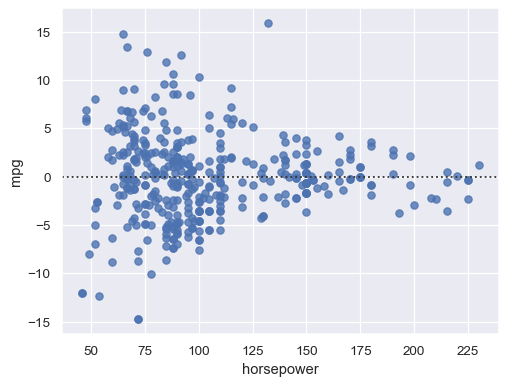
Remove higher-order trends to test whether that stabilizes the residuals:
sns.residplot(data=mpg, x="horsepower", y="mpg", order=2)
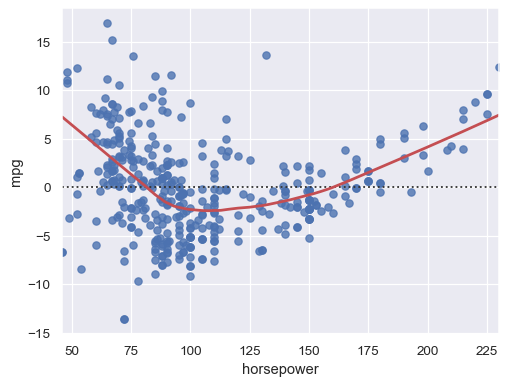
Adding a LOWESS curve can help reveal or emphasize structure:
sns.residplot(data=mpg, x="horsepower", y="mpg", lowess=True, line_kws=dict(color="r"))