Genetic Network Identification using Convex Programming
IET Systems Biology, 3(3):155-166, May 2009.
Shorter version appeared as
Proceedings 8th International Conference on Systems Biology,
paper F15, October 2007.
Gene regulatory networks capture interactions between genes and other cell
substances, resulting in various models for the fundamental biological process
of transcription and translation. The expression levels of the genes are
typically measured as mRNA concentration in micro-array experiments. In a
so-called genetic perturbation experiment, small perturbations are applied to
equilibrium states and the resulting changes in expression activity are
measured. One of the most important problems in systems biology is to use these
data to identify the interaction pattern between genes in a regulatory network,
especially in a large scale network. The authors develop a novel algorithm
for identifying the smallest genetic network that explains genetic
perturbation experimental data. By construction, our identification algorithm
is able to incorporate and respect a priori knowledge
known about the network structure. A priori biological knowledge is
typically qualitative, encoding whether one gene affects another gene or not,
or whether the effect is positive or negative. The method is based on a
convex programming relaxation of the combinatorially hard problem of 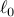 minimisation. The authors apply the proposed method to the identification
of a subnet work of the SOS pathway in Escherichia coli, the segmentation
polarity network in Drosophila melanogaster, and an artificial network for
measuring the performance of the method.
minimisation. The authors apply the proposed method to the identification
of a subnet work of the SOS pathway in Escherichia coli, the segmentation
polarity network in Drosophila melanogaster, and an artificial network for
measuring the performance of the method.