Replication of the Virion and Molecular Biology
There are two stages of the replication cycle of adenoviruses: early and late. The virus first attaches to the host cell via the fibers, which bind to specific receptors on the host cell plasma membrane. The virion then enters the cell via endocytosis in a clathrin-coated vesicle. The pentons are removed due to a pH drop within the vesicle, and a spherical shaped capsid is released. This capsid travels to the nucleus via microtubules, where the genome then enters the nucleus via nuclear pores. Witihin only a couple hours, the viral genome shuts down DNA and protein synthesis of the host. The viral DNA is incorporated into a complex with cellular histones, and is then able to bind to the nuclear matrix and proceed with replication.
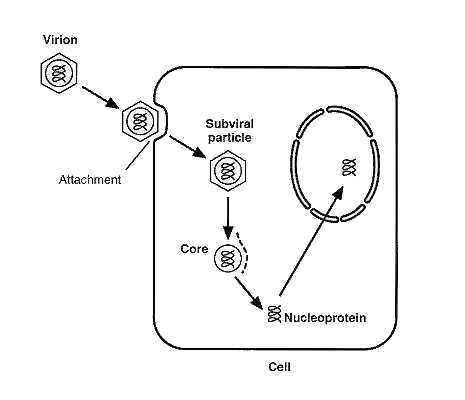
courtesy of: Walter Doerfler
The proteins of adenoviruses are generated in a highly ordered and controlled fashion. There are essentially three phases: immediate early, early, and late. There is one immediate early mRNA transcript, called E1A. This mRNA is translated to yield a protein which is important for transcriptional activation of early genes.
In the early phase, mRNA transcripts for E1B, E2A, E2B, E3, E4, and some structural proteins (II, III, IIIa, etc) are generated. E1A and E1B are essential for viral replication, and together are known to be sufficient to cause transformation in rodent cells. This discovery was the first time a virus was shown to have oncogenic potential. In more detail, E1A has been shown to cooperate with ras (small G protein, important in an enormous number of cellular functions), and more recently, Rb105 (tumor-supressor gene). Meanwhile, E1B has been shown to cooperate with myc, as well as p53 (another tumor-supressor gene). The functions of the other transcripts are as follows: E2A translates a DNA binding protein (DBP) which regulates viral DNA replication (discussed below) as well as regulating expression of late gene, E2B encodes a DNA polymerase, and (due to a post-translational modification) a precursor to the terminal protein (pTP a 55kd protein covalently linked to the 5' end of both strands of the genome which catalyzes viral DNA replication). The function of E3 is unknown, and is not essential for tissue culture infection. E4 is a protein used in immune modulation.
After the immediate early and early proteins are translated, DNA replication begins. Essentially, this process involves 3 proteins: The 5' associated terminal protein described about (pTP) acts as a primer of initiation for both strands. There is also a DNA binding protein (E2A) and the polymerase (E2B).
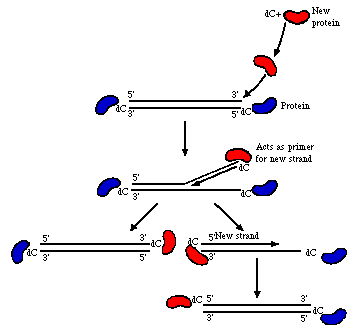
courtesy of Dr. Alan Cann; also in Fields, p.1699
Cellular proteins, such as NFI, NFII, topoisomerase I, also participate in this process. One proposed mechanism involves circularization of the genome. This is shown below:
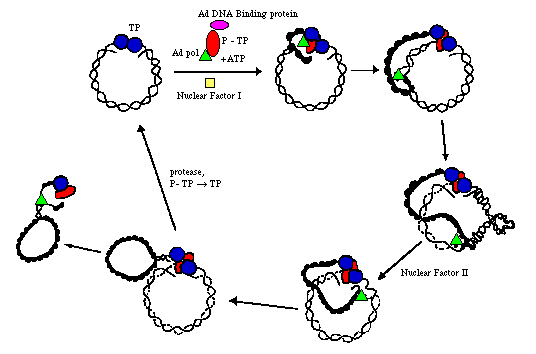
courtesy of Dr. Alan Cann; also in Fields, p.1702
Around the same time, there is a switch from early to late genes which is poorly understood, but highly regulated by the virus. The late genes encode a series of virion proteins which are necessary for assembly and maturation. Interestingly, the initiation is controlled by a single promoter, yielding a single primary RNA transcript. This tripartite leader (3 exons) transcript is differentially spliced by a highly studied mechanism to yield at least 13 secondary mRNA transcripts. For an unknown reason, only newly synthesized DNA is used as templates for the production of late genes. This may be a regulatory process. That is, the structural proteins are not made until enough DNA has been made to fill them (This is only my personal hand-waving!). In any case, empty capsid shells are assembled in the cytoplasm while the core proteins and genome are assembled in the nucleus. Eosinophilic crystals (inclusion bodies) accumulate in the nucleus, just prior to lysis of the cell. At this point, the nuclear membrane is broken down and the genomic core and proteins are encased within the capsid proteins. This enormous amplification ultimately results in release of the virions by cellular damage and/or lysis.


