Final Project Proposals
Analyzing Connectomic Networks
The landing page for the Network Data Analysis group at Ohio State University can be found here. If you want to get up to speed on their approach as outlined in Chowdhury and Memoli [7], Peter Li suggested the following progression: one, two, three and four. The first three just develop and apply the Dowker Complex as a simplicial complex sensitive to asymmetry, but without a homological operator that retains asymmetries, so the sensitivity of the overall persistence diagrams to asymmetry is modest.
The last paper—four—makes the additional step of introducing ordered simplices and ordered homology, so the resulting persistence strongly reflects asymmetries. Pawel Dlotko notes that this is computationally expensive, and Peter Li1 has concerns about its practicality and is not sure how good the available software support is. If you are already familiar with persistence over simplicial complexes from presentation by Pawel Dlotko2 last year (VIDEO), you can probably jump right into the last paper, but you may find it helpful to go through all of them just to get multiple restatements of the same information as well as to understand the progression. Both Peter and Pawel would be happy to help out teams working on this topic.
Sebastian Seung's Presentation
Papers on Seung and Zung's Correlation Game plus two of Peter Földiák's papers: [Seung and Zung], [Carlson], [Földiák], [Földiák]
Drosophila Structure and Function
Papers on Drosophila Structure and Function — click on the item for the PDF: Neriec and Desplan, Plaza et al, Takemura et al, Borst and Euler, Spalthoff, Joesch et al, Erclik et al, Mauss et al, Borst, Bahl et al, Maisak et al, Reiff et al, Borst and Helmstaedter, Behnia et al, Morante and Desplan. Art Pope3 would be happy to help out teams working on applying graph search and computer vision algorithms to the Janelia data.
the Here are a few visual aids selected from the these papers:
Borst and Helmstaedter [5] Figure 4:
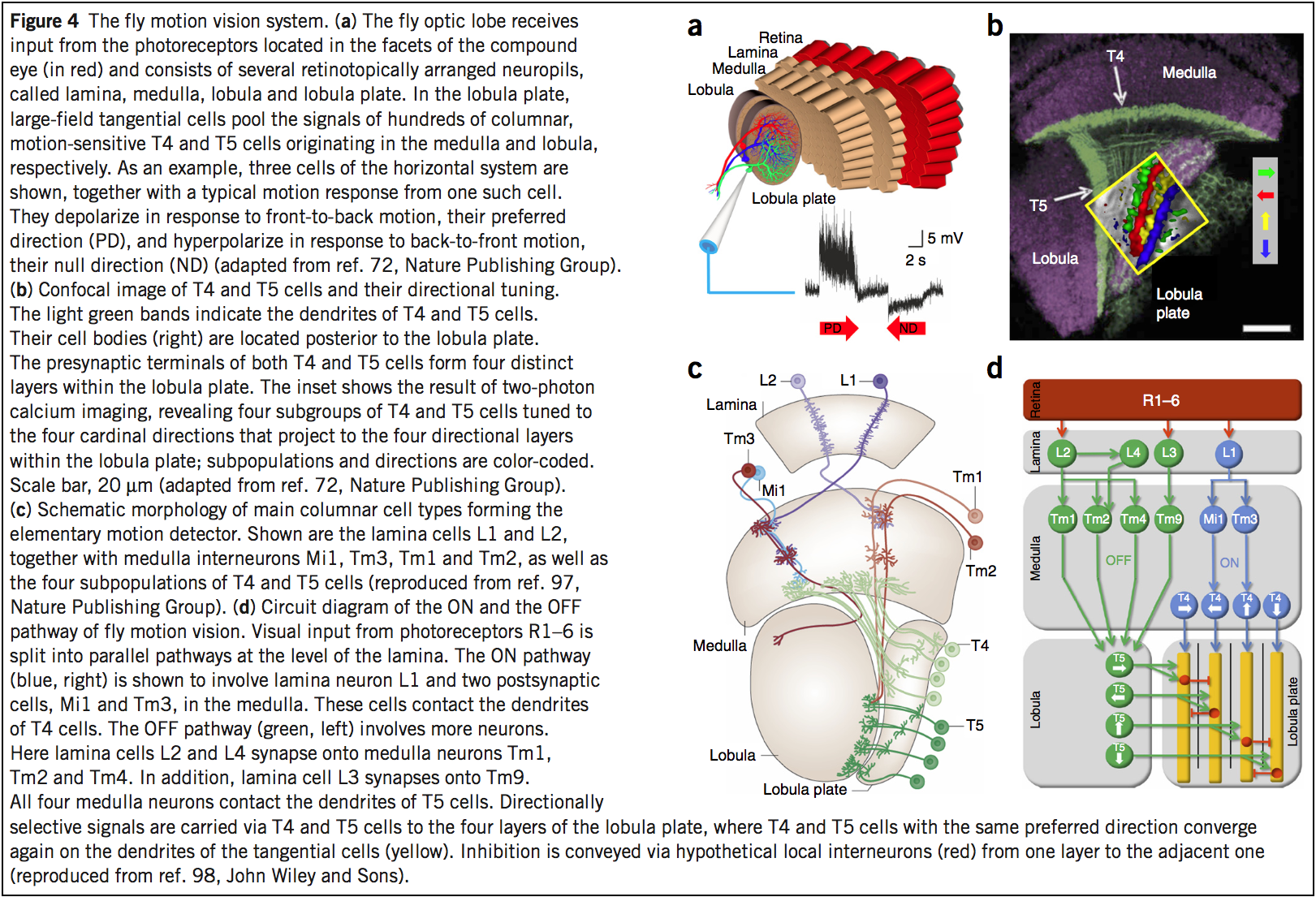 |
Behnia et al [2] Figure 1:
 |
Morante and Desplan [15] Figure 2:
 |
Seven Column Medulla Dataset
The Python code for exploring the seven-column dataset and analyzing the data using ideas from algebraic topology can be found here: [NEUROBOT] [CODE]. There are 15 neurons of type, Mi1, in the seven-column dataset of which only seven — Mi1-H (center column) and Mi1-{A-F} (adjoining six column) — are relevant. The remaining eight are likely only partially reconstructed. Aside from the skeleton files — found in ./code/github/FlyConnectome/skeletons/, the primary sources of data in the ./code/github/FlyConnectome/ respository consist of two json files containing Python dict structures:
neuronsinfo.json — everything you need to know about 462 fully-traced neurons and over 5,000 partially-traced neurons;
synapses.json — everything you need to know about 53,383 synapses organized in small cohorts called T-bar structures;
References
1 Peter Li is a neuroscientist working on the Neuromancer Team at Google and focusing on structural connectomics. Peter did his Ph.D. with E.J. Chichilnisky at the Salk Institute before E.J. moved Stanford.
2 Pawel Dlotko is a mathematician working in the area algebraic topology. After finishing a postdoc at INRIA last year, Pawel and his family are now living in the UK where he teaches and conducts research.
3 Art Pope is a software engineer and expert in computer vision. Art works on the Neuromancer team specializing on aligning, registering and stitching together very-large 3D volumes of electronmicroscopy data.